- Title
-
Metabolic reprogramming and membrane glycan remodeling as potential drivers of zebrafish heart regeneration
- Authors
- Spelat, R., Ferro, F., Contessotto, P., Aljaabary, A., Martin-Saldaña, S., Jin, C., Karlsson, N.G., Grealy, M., Hilscher, M.M., Magni, F., Chinello, C., Kilcoyne, M., Pandit, A.
- Source
- Full text @ Commun Biol
|
The response to injury is characterized by molecular and metabolic changes.
a GOCircle plot summarizing gene ontology enrichment analysis at 2 dpci, b 7 dpci and c 14 dpci; “carbohydrate metabolic process” and “N-linked glycosylation” are identified as significantly activated processes, among others. The significance of each term (-log10 adjusted p value) is specified by the height of the bar plot in the inner ring, while the color corresponds to the z-score. The expression level (log2FC) for the genes in each term is displayed in the outer ring scatterplots; red dots indicated upregulated genes and blue dots downregulated genes. d The most statistically significant canonical pathways for the transcriptomic analysis at 2 dpci, e 7 dpci and f 14 dpci identified using the IPA® software are listed according to their p value (-log); the threshold -log(p value) = 1.3 corresponds to a p value of 0.05. Activation is indicated in red, inhibition in green and unpredictability in gray. g The most statistically significant canonical pathways for the proteomic analysis at 2 dpci, h 7 dpci and i 14 dpci. j Heatmap showing glycolytic enzymes and mitochondrial oxidative phosphorylation proteins at 2 dpci, k 7 dpci and l 14 dpci. Two biological replicates deriving from the pooling of 4 different animals (n = 8 animals per group) were analyzed for all experiments. For proteomic analysis n = 4 animals per group were used. |
|
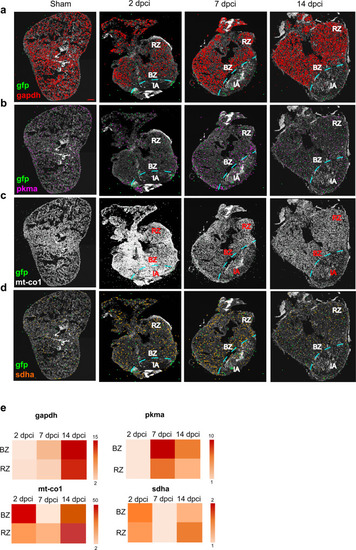
Cardiomyocyte targeted in situ sequencing.
A targeted multiplexed mRNA detection assay employing padlock probes, rolling circle amplification (RCA), and barcode sequencing was applied to detect the spatial distribution of a gapdh b pkma c mt-co1 and d sdha in sham and at 2, 7 and 14 dpci. All the experiments were performed on cmlc2 zebrafish transgenic strain, where a green fluorescent protein (gfp) gene was inserted under the promoter of cardiomyocyte-specific gene cardiac myosin light chain 2. For this reason, gfp was chosen as a cardiomyocyte marker. IA injured area, BZ border zone, RZ remote zone. Scale bar = 100 μm. e Heatmaps showing the expression comparison of gapdh, pkma, mt-co1 and sdha between BZ and RZ at each time point. Heat legend represents the relative quantification of the spot number and it is represented as fold change. Two biological replicates were analyzed. |
|
Lectin microarray analysis evidences cell membrane sialylation alterations in zebrafish heart during regeneration.
a Hierarchical clustering of lectin microarray data. Asialofetuin (ASF) was used as control glycoprotein and all lectin binding intensities were normalized. b Lectin microarray dot plot of 2 dpci, c 7 dpci and d 14 dpci in respect to the sham showing a reduction of: sialylation (MAA, WGA), fucosylation (LTA, UEA-I), GlcNAc (WGA) and Gal/GalNAc (RPbAI) at 2 dpci; sialylation (MAA), fucosylation (LTA, UEA-I), mannose (Con A, Lch-A), Gal/GalNAc (AIA) and GalNAc (SBA) at 7 dpci; and Gal/GalNAc (RPbAI) at 14 dpci. One-way ANOVA analysis with Tukey’s post hoc correction was applied. Spot colors correspond to lectin sugar specificity; the red dotted line represents a significant cutoff of p value ≤ 0.05. Three biological replicates were analyzed, each one deriving from the pooling of 4 different animals (n = 12). |
|
Sialic acid distribution in regenerating zebrafish heart identified by lectin histochemistry.
a Lectin histochemistry showing the distribution of MAA, recognizing α-(2,3)-linked sialic acid, b SNA-I, binding to α-(2,6)-linked sialic acid and c WGA, binding to sialic acid and GlcNAc, in ventricle tissue. d MAA, e SNA-I and f WGA relative quantification. The lectin binding quantifications highlighted a similar decrease at 2 dpci and a subsequent increase in the following timepoints. Cardiomyocytes were identified by GFP positivity. IA injured area. Data are shown as mean with SD (n = 4 animals per group). Scale bar = 100 μm. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. |
|
Membrane N-glycan profile of regenerating zebrafish heart established by LC-MS/MS revealed increased sialylation and a reduction in high mannose structures during the inflammatory phase.
a Comparison of N-glycan types (complex, high mannose and hybrid) and sialylation between sham and the different regeneration phases (2, 7 and 14 dpci). The inflammatory phase (2 dpci) showed a 6% increase in sialylated glycans and a 5% decrease in high mannose structures compared to sham. b Summary of the main changes in N-glycans features (sialylated hybrid and complex type increase/decrease, non-sialylated hydrid and complex type increase/decrease, high mannose-type increase/decrease) showing some of the corresponding structures. c Heatmap displaying time-specific relative quantification of neutral, and d sialylated N-glycans. Color intensity represents the relative abundance expressed as a percentage. e Extracted ion chromatography (EIC) showing the most abundant membrane N-glycans present in zebrafish heart. Results deriving from samples pooling of six animals are shown (n = 6 animals per group). |
|
Membrane O-glycan profile of regenerating zebrafish heart established by LC-MS/MS revealed decreased sialylation during the inflammatory phase.
a Comparison of O-glycan structures (core 1 and core 2) and sialylation between sham and the different regeneration phases (2, 7 and 14 dpci). The inflammatory phase (2 dpci) showed a 12% decrease in sialylated O-glycans and a subsequent increase at 7 and 14 dpci. b Summary of the main changes in O-glycan structures (sialylated core 1/2 increase/decrease, non-sialylated core 1/2 increase/decrease) showing some of the corresponding structures. c Heatmap displaying time-specific relative quantification of core 1 and core 2 O-glycans. Sialylated structures are also indicated (purple square). Color intensity represents the relative abundance expressed as a percentage. d EIC showing the most abundant membrane O-glycans present in zebrafish heart. Results deriving from samples from the pooling of six animals are shown (n = 6 animals per group). |