- Title
-
Zebrafish as a Vertebrate Model for Studying Nodavirus Infections
- Authors
- Lama, R., Pereiro, P., Figueras, A., Novoa, B.
- Source
- Full text @ Front Immunol
|
Survival rates of zebrafish larvae challenged with NNV through different infection routes and NNV replication. |
|
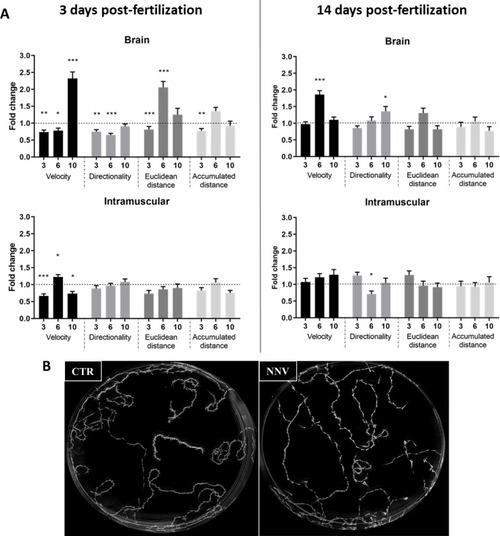
Image analysis of the swimming behavior of zebrafish larvae (3 and 14 dpf) infected with NNV |
|
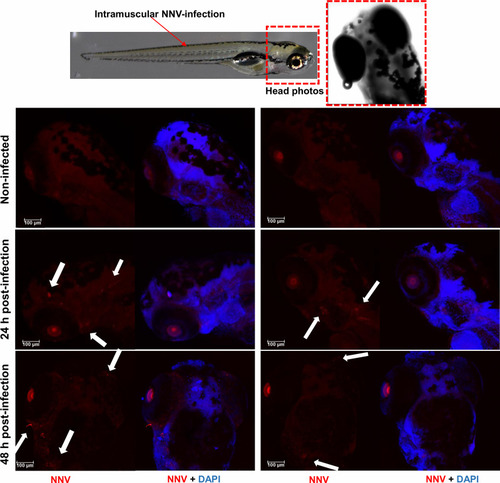
Whole-mount immunofluorescence of zebrafish larvae infected by intramuscular microinjection with NNV. Confocal images of the head from uninfected and NNV-infected larvae at 24 and 48 hpi. NNV particles are stained red, and cell nuclei are stained blue (DAPI). White arrows denote the position of NNV-infected cells. |
|
Visualization and analysis of innate immune cell migration to the head of 3 pdf larvae infected through different routes. The transgenic zebrafish lines |
|
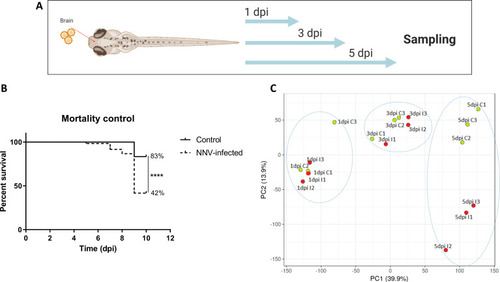
Transcriptome analysis of 3-dpf zebrafish larvae infected with NNV |
|
Differentially expressed genes in zebrafish larvae infected with NNV. |
|
GO terms and KEGG pathways enriched during NNV infection of zebrafish larvae. |
|
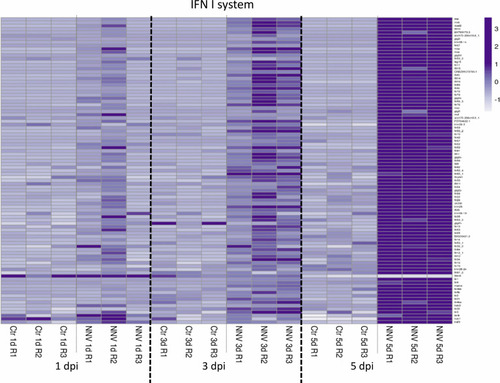
Heatmap representing the DEGs linked to the type I IFN system at 1, 3, and 5 dpi. A heatmap was constructed with the TPM expression values of the DEGs. Expression levels are represented as row-normalized values on a white–purple colour scale. |
|
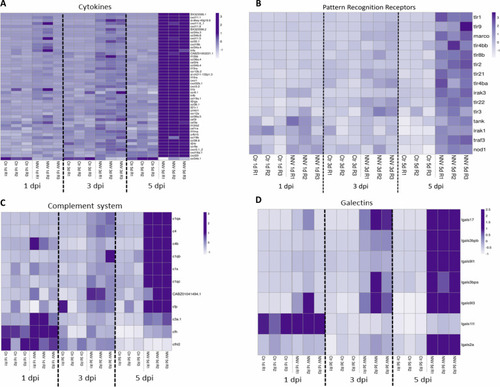
Heatmaps representing the DEGs belonging to different immune categories at 1, 3 and 5 dpi: |