- Title
-
Differential electrographic signatures generated by mechanistically-diverse seizurogenic compounds in the larval zebrafish brain
- Authors
- Pinion, J., Walsh, C., Goodfellow, M., Randall, A.D., Tyler, C.R., Winter, M.J.
- Source
- Full text @ eNeuro
|
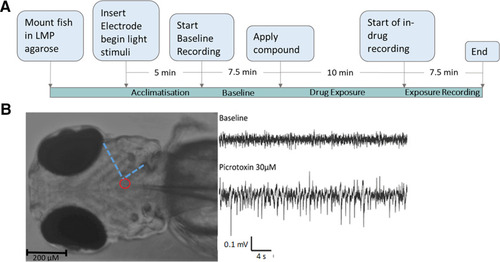
A schematic showing the experimental process and example data from |
|
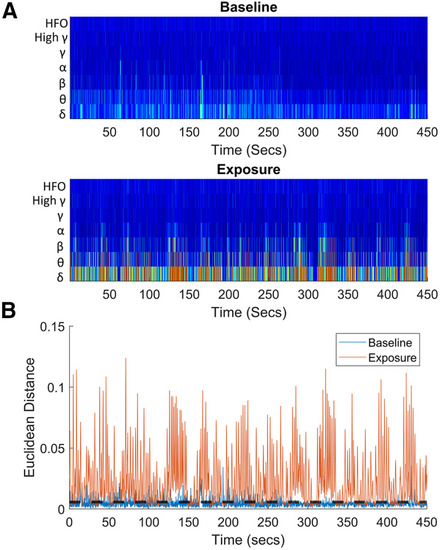
Example data obtained from |
|
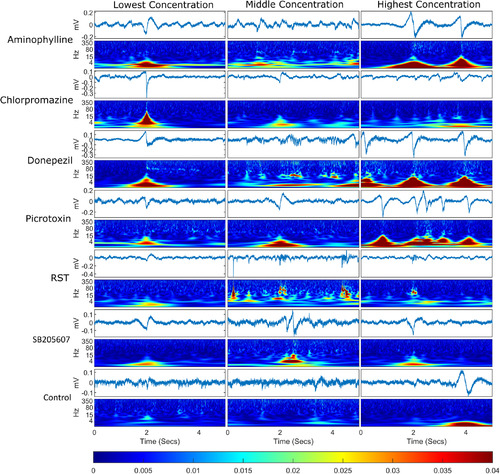
Example of an event plus 2 s either side for each treatment group. For each treatment group, the timeseries are displayed for each event with a wavelet transform below each one displaying the frequency domain over the same time period. The events selected were the events whose spectra were closest in Euclidean distance to the mean event spectra for that treatment group, meaning that these are representative of the events shown for each compound. The bottom bar shows the color scaling for the magnitude of the wavelet transformation. Each column represents a different concentration set and each row represents a different compound. |
|
Mean number of events detected per treatment group. Bar graph showing the mean number of events per treatment group. Error bars represent the SEM ( |
|
Analysis of the AUC of the Hilbert transform of the LFP recordings. |
|
Data generated for larvae exposed to each of the test compounds after spectral analysis and categorization into specific frequency bands. |
|
Multi-dimensional scaling (MDS) of the normalized power spectra. Left-hand image, A scatter graph of the first two coordinates produced after classical MDS of each individual larva. In both plots, the circles represent the lowest concentration, the triangles the middle concentration, and the diamonds the highest concentration used for each treatment group. The black square represents the control group. Right-hand image, A scatter graph of the first two coordinates produced after classical MDS of the mean normalized power spectra of each treatment group (see |