- Title
-
Construction and Evaluation of Recombinant Attenuated Edwardsiella piscicida Vaccine (RAEV) Vector System Encoding Ichthyophthirius multifiliis (Ich) Antigen IAG52B
- Authors
- Swain, B., Powell, C.T., Curtiss, R.
- Source
- Full text @ Front Immunol
|
Phylogenetic and structural similarity of |
|
|
|
Complementation of Δ |
|
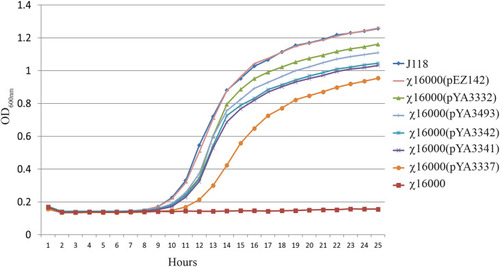
Growth of |
|
IAG52B sequence analysis. |
|
|
|
Dissemination and colonization of χ16022(pG8R8029) in zebrafish tissues. |
|
|
|
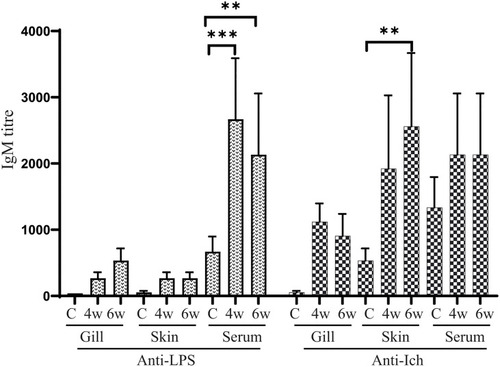
Anti-LPS and anti-Ich antibody responses in zebrafish. Serum and mucosal immunoglobulin M (IgM) responses to |
|
Survival of χ16022(pG8R8029), χ16022(pYA3493) vaccinated and BSG control fish after wild-type E. piscicida challenge. Control and 4 weeks post-vaccination zebrafish were i.c. challenged with 1 × 105 CFU/dose (10 × LD50) of wild-type |