- Title
-
Loss of IRF2BPL impairs neuronal maintenance through excess Wnt signaling
- Authors
- Marcogliese, P.C., Dutta, D., Ray, S.S., Dang, N.D.P., Zuo, Z., Wang, Y., Lu, D., Fazal, F., Ravenscroft, T.A., Chung, H., Kanca, O., Wan, J., Douine, E.D., Network, U.D., Pena, L.D.M., Yamamoto, S., Nelson, S.F., Might, M., Meyer, K.C., Yeo, N.C., Bellen, H.J.
- Source
- Full text @ Sci Adv
|
IRF2BPL overexpression decreases wg transcript, protein, and signaling in the wing disc.
(A) Schematic of the developing wing pouch where the Wg expression along the DV boundary develops into the adult wing margin. Optical images of the adult fly wing upon overexpression of UAS-LacZ or UAS-IRF2BPL constructs with nub-GAL4 at 18°C. (B) Wnt pathway in the developing wing pouch at the DV boundary. (C and D) Third instar larval wing disc upon overexpression of UAS-LacZ or UAS-IRF2BPL with nub-GAL4 at 18°C and stained with anti-Wg or Wnt downstream targets anti-Cut and anti-Sens (C) or the wg-lacZ reporter gene (D). Note the absence of Wg or its downstream responders by the arrows. (E) IRF2BPL protein structure and schematic of deletion constructs. (F) Third instar larval wing disc upon overexpression of UAS-IRF2BPL constructs with nub-GAL4 at 18°C and stained with anti-Wg or Wnt downstream targets anti-Cut and anti-Sens. Scale bars, 40 μm. |
|
Neuronal reduction of Pits reduces life span and causes progressive climbing defects and axonal loss.
(A) Schematic of Pits locus in Drosophila with original PitsMI02926 insertion that was converted to Pits::GFPDH by injection of Double Header plasmid (20), resulting in an internally tagged protein trap allele that affects all known Pits transcripts. (B) GFP-tagged fusion proteins can be targeted by deGradFP (UAS-NSlmb::vhhGFP4) to degrade Pits::GFPDH with spatial (GAL4) and temporal (temperature) control. (C and D) Life span is decreased when Pits::GFPDH is ubiquitously (C) or neuronally (D) degraded by deGradFP during development and the adult stage. n is a minimum of 48 flies per genotype (****P < 0.0001). Statistical analyses were determined by the log-rank test. (E) Pits::GFPDH; nSyb-GAL4, UAS-deGradFP flies exhibit progressive climbing defects. Time (seconds) required for flies of the indicated genotypes to climb past 7 cm (n > 20 per genotype). Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are means ± SEM (****P < 0.0001). (F and H) Diagram of the peripheral wing nerves containing peripheral neurons of the anterior wing margin observed with (G and H) transmission electron microscopy. Quantification shows the total axon number at (G) 5 d.p.e. in Pits::GFPDH; nSyb-GAL4, UAS-deGradFP flies containing GR construct (n = 3) and flies without GR (n = 4) and (H) 25 d.p.e. (n = 5 each genotype). Results are means ± SEM (***P < 0.001). Statistical analyses were determined by two-tailed Student’s t test. Scale bars, 1 μm. |
|
Wingless genetically interacts with IRF2BPL, and neuronal reduction of Pits in neurons leads to increased wg transcription.
(A and B) Coexpression of UAS-IRF2BPL with UAS-Wg::HA, but not UAS-LacZ, in the wing disc using nub-GAL4 at 18°C restores Wg expression along the DV boundary (A) and the adult wing margin morphology (B). Scale bar, 40 μm. (C to F) Neuronal reduction of Pits via RNAi using elav-GAL4, UAS-dicer results in increased wg transcript (C) by qPCR and Wg protein (D) in 20 d.p.e. fly heads, as quantified in (E) and by immunohistochemistry (F) in the adult brain of 20-day-old flies stained for Wg. Results are means ± SEM (**P < 0.01). Statistical analyses were determined by two-tailed Student’s t test. Scale bar, 50 μm. (G) Climbing assessment of Pits::GFPDH; nSyb-GAL4, UAS-deGradFP flies at 25 d.p.e. after adult-specific reduction of Pits::GFP and treatment with DMSO or indicated Wnt inhibitors. Time (seconds) required for flies of the indicated genotypes to climb past 7 cm (n > 20 per genotype). Statistical analyses were determined by ANOVA followed by Tukey post hoc. Results are means ± SEM (**P < 0.01 and ***P < 0.001). |
|
Neuronal expression of Wg ligand is sufficient to cause progressive climbing defects and axonal loss.
(A) Survival curves for neuronal expression of UAS-LacZ or UAS-Wg::HA using nSyb-GAL4 at 25°C. A minimum of 43 flies per genotype (*P < 0.05). Statistical analyses were determined by the log-rank test. (B) nSyb-GAL4, UAS-Wg::HA flies exhibit progressive climbing defects. Time (seconds) required for flies of the indicated genotypes to climb past 7 cm (n > 20 per genotype). Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are means ± SEM (**P < 0.01). (C) nSyb-GAL4, UAS-Wg::HA flies exhibit flight defects at 25 d.p.e. compared to nSyb-GAL4, UAS-LacZ controls when dropped into the center of a 2-liter graduated cylinder (n > 32 per genotype). Results are means ± SEM (**P < 0.01). Statistical analyses were determined by two-tailed Student’s t test. (D and E) Transmission electron microscopy of the peripheral wing nerves containing peripheral neurons of the anterior wing margin. Quantification shows the total axon number at (E) 5 d.p.e. upon neuronal (nSyb-GAL4) overexpression of UAS-LacZ or UAS-Wg::HA (n = 5 each genotype) and the same genotypes at 25 d.p.e. (UAS-LacZ, n = 4; UAS-Wg::HA, n = 5). Results are means ± SEM (**P < 0.01). Statistical analyses were determined by two-tailed Student’s t test. Scale bars, 1 μm. |
|
Loss of irf2bpl in zebrafish leads to defective locomotor activity and increased Wnt1 signaling.
(A) Distance moved (in millimeters) of mutants (irf2bpl−/−) and wild-type sibling controls (irf2bpl+/+) measured at 7 d.p.f. in a locomotor assay. During the assay, test animals were first habituated for 30 min, followed by five light-dark cycles for a total of 30 min (n = 36 and 47 for irf2bpl+/+and irf2bpl−/− animals, respectively). (B) Representative Western blot and quantification of Wnt1 protein expression in irf2bpl−/− brain compared to irf2bpl+/+ controls (n = 3 per group). (C) Analyses of lef1 transcript expression real-time qPCR in irf2bpl−/− treated with DMSO, SSTC3, or XAV-939, compared to irf2bpl+/+ treated with DMSO (n = 8 per group). (D and E) Treatment with SSTC3 significantly improved the irf2bpl−/− mutants’ bout activity when transitioning from a light to dark condition, compared to mutants treated with DMSO (n = 22 and 24 for DMSO-treated irf2bpl+/+ and irf2bpl−/− animals; 25 and 23 for SSTC3-treated irf2bpl+/+ and irf2bpl−/−, respectively). (F and G) Similar to SSTC3, treatment with XAV-939 significantly improved the mutants’ bout activity compared to DMSO-treated mutants (n = 21 and 19 for DMSO-treated irf2bpl+/+ and irf2bpl−/− animals; 36 and 45 for XAV-939–treated irf2bpl+/+ and irf2bpl−/− animals, respectively). Results are means ± SEM (*P < 0.05, **P < 0.001, and ****P < 0.0001). |
|
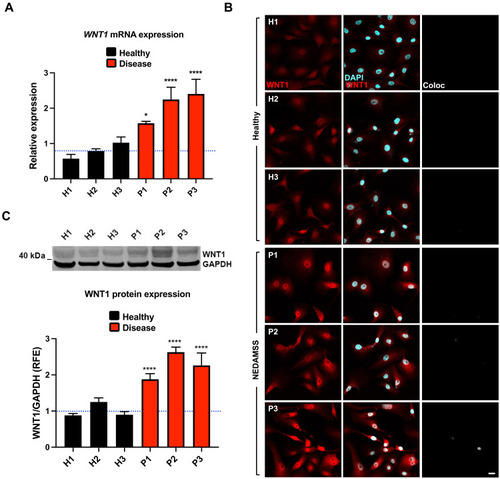
Reprogrammed astrocytes from NEDAMSS patients display increased WNT1 transcription and protein.
(A to C) WNT1 transcript (A) and protein (B and C) are elevated in reprogrammed astrocytes derived from fibroblasts of NEDAMSS patients and healthy controls by qPCR (A), immunofluorescence (B), and Western blot (C). RFE, relative fold expression. ANOVA followed by Dunnett’s multiple comparison test between the mean of the controls and the mean of each line was computed to derive the P value: *P < 0.05 and ****P < 0.0001. The experiments were conducted three times. Scale bar, 20 μm. |
|
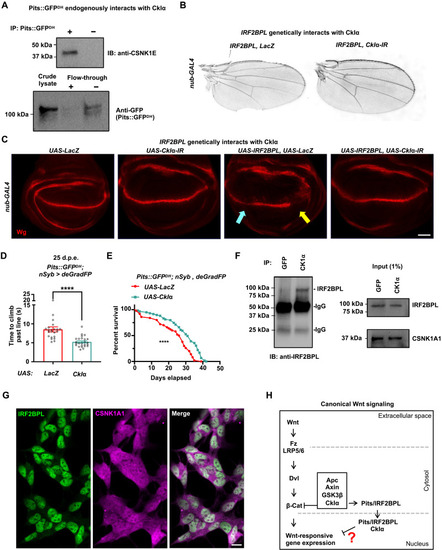
Pits physically and genetically interacts with CkIα.
(A) Pulldown of Pits::GFPDH from fly heads indicates that endogenous CkIα interacts with Pits detected by Western blotting using anti-CSNK1E antibody (CkIα size is ~38 kDa). Interaction observed in triplicate. (B and C) Coexpression of UAS-IRF2BPL with UAS-CkIα-IR, but not UAS-LacZ, in the wing disc using nub-GAL4 at 18°C restores adult wing margin morphology (B) and Wg expression along the DV boundary (C). Scale bar, 40 μm. (D) Pits::GFPDH; nSyb-GAL4, UAS-deGradFP coexpressing either UAS-LacZ or UAS-CkIα::HA. Time (seconds) required for flies of the indicated genotypes to climb past 7 cm (n > 22 per genotype). Statistical analyses were determined by two-tailed Student’s t test. (E) Survival curves for Pits::GFPDH; nSyb-GAL4, UAS-deGradFP coexpressing either UAS-LacZ or UAS-CkIα::HA. n is a minimum of 49 flies per genotype (****P < 0.0001). Statistical analyses were determined by the log-rank test. (F) In vitro coimmunoprecipitation of CKIα (CSNK1A1) or GFP as a control in SH-SY5Y cells showing an interaction with endogenous IRF2BPL. IgG, immunoglobulin G. (G) SH-SY5Y cells stained with IRF2BPL and CSNK1A1. Scale bar, 10 μm. (H) Schematic depicting the canonical Wnt pathway, highlighting potential actions of Pits/IRF2BPL. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |