- Title
-
Loss of zebrafish atp6v1e1b, encoding a subunit of vacuolar ATPase, recapitulates human ARCL type 2C syndrome and identifies multiple pathobiological signatures
- Authors
- Pottie, L., Van Gool, W., Vanhooydonck, M., Hanisch, F.G., Goeminne, G., Rajkovic, A., Coucke, P., Sips, P., Callewaert, B.
- Source
- Full text @ PLoS Genet.
|
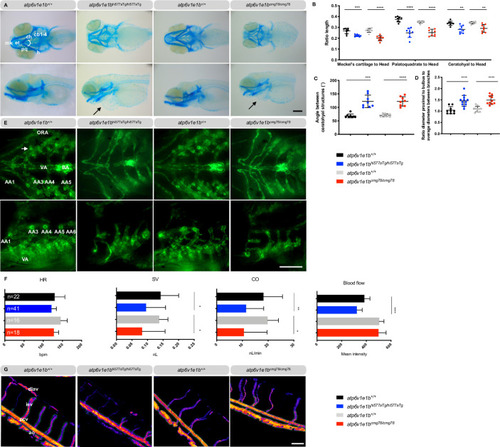
Representative images are shown. (A) Ventral (top panel) and lateral views (bottom panel) of Alcian blue stained craniofacial structures at 5 dpf reveal misshapen and shorter Meckel’s cartilage (m), shorter palatoquadrate (pq) and shorter ceratohyal (ch) structure and a higher angle between ch structures in |
|
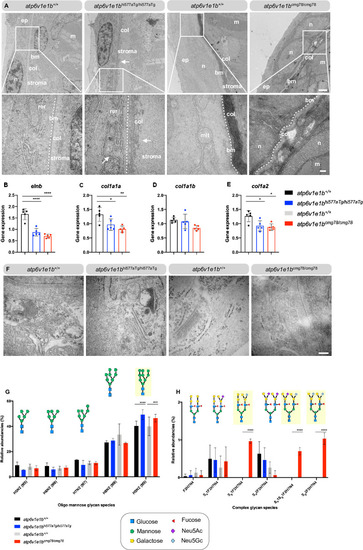
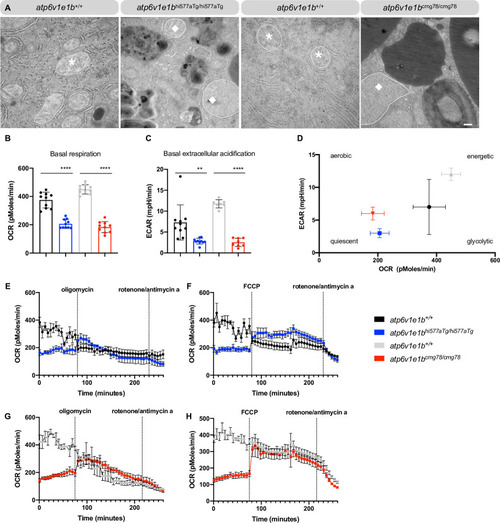
(A) Representative images of ultrathin sections taken from the dermis of |
|
(A) Phosphorylated ribosomal protein S6 kinase 1 (S6K1) (upper panel) and total S6K1 (lower panel) were detected in protein lysates of whole |
|
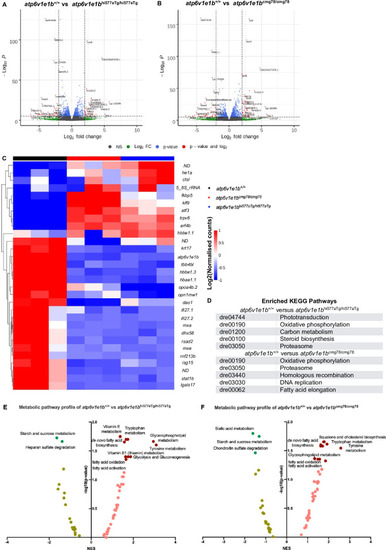
(A) Volcano plot of RNA-sequencing data of |
|
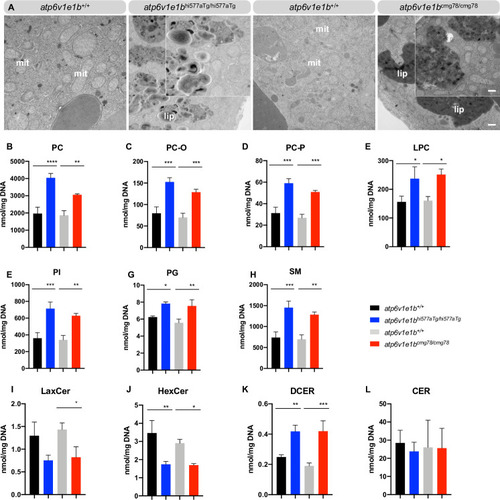
(A) Representative images of ultrathin sections of the yolk from 4 dpf WT control and |
|
(A) Representative images of ultrathin sections of the yolk from WT control and |