- Title
-
Ccdc103 promotes myeloid cell proliferation and migration independent of motile cilia
- Authors
- Falkenberg, L.G., Beckman, S.A., Ravisankar, P., Dohn, T.E., Waxman, J.S.
- Source
- Full text @ Dis. Model. Mech.
|
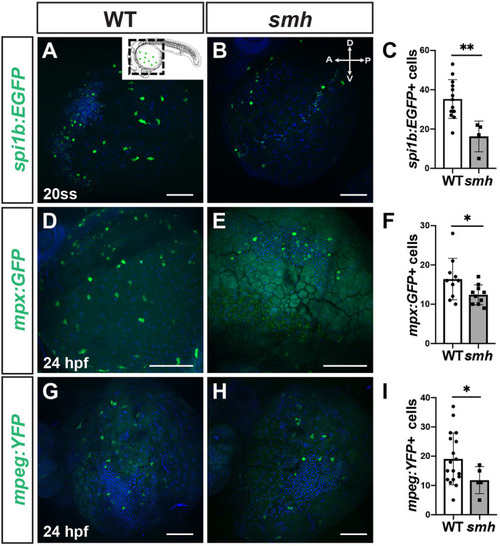
EXPRESSION / LABELING:
|
|
|
|
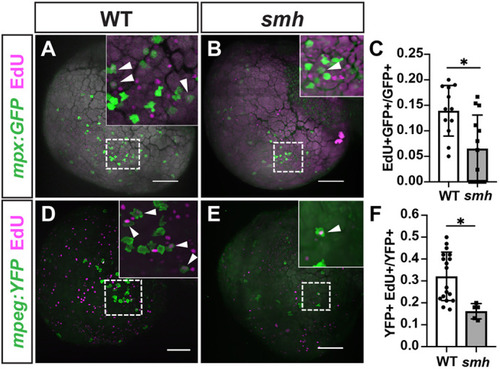
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
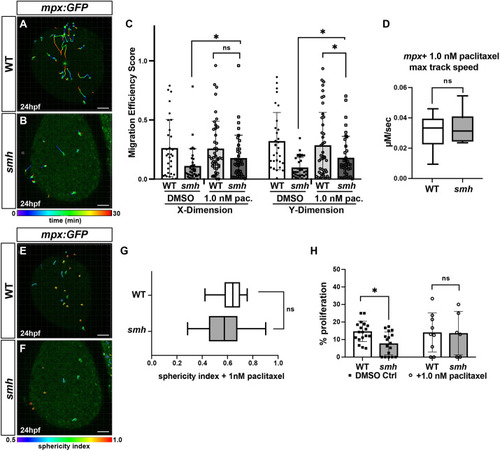
(A,B) Representative confocal projection images from wounding experiments of paclitaxel-treated WT (n=4) and smh (n=4) mpx:EGFP transgenic embryos at 24 hpf. (C) Quantification of migration efficiency scores calculated from point position data generated in Imaris. Each data point represents an individual cell from a minimum of three separate experiments, per genotype, per treatment. (D) Quantification of maximum track speed. Each data point represents an individual cell. Cell tracks and sphericity were generated by Imaris. (E,F) Maximum confocal projections of wounded, paclitaxel-treated WT and smh embryos bearing the mpx:GFP transgene with individual cells projected as 3D surfaces and color coded according to sphericity index as calculated by Imaris. Red, more spherical; blue, less spherical. (G) Mean cell sphericity indices as calculated in Imaris. In D and G, the box and whisker plots represent the median value (center line), with the box including all values from the two median quartiles and the whiskers representing minimum and maximum values (with no outliers excluded). (H) The percentage of EdU+/mpx:EGFP+ cells from transgenic WT and smh mutant embryos, pulsed with EdU at 17-ss and treated with paclitaxel from 17-ss to 24 hpf. *P<0.05; ns, not significant (two-tailed unpaired Student's t-test). Scale bars: 100 μm. PHENOTYPE:
|
|
|
|
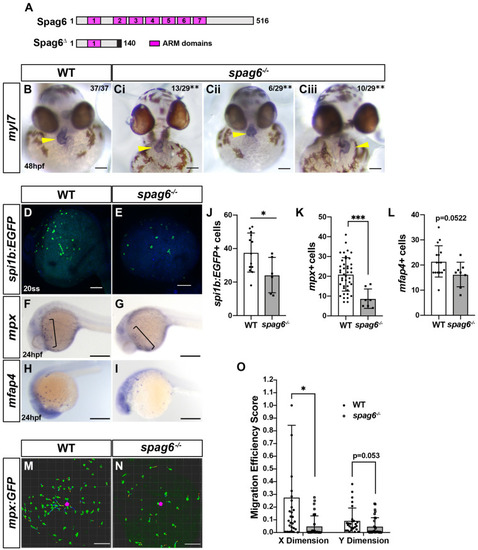
(A) Domain architecture for WT zebrafish Spag6 and the predicted truncation from the spag6 mutant allele used. (B,Ci-iii) ISH for myl7 in WT and spag6 mutant embryos at 48 hpf. (B,Ci) Normally (situs solitus, dextral) looped hearts in WT and spag6 mutant embryos. (Cii) Linearized heart in spag6 mutant. (Ciii) Reverse (situs inversus) heart in spag6 mutant. Yellow arrowheads indicate hearts. Fractions indicate the number of embryos with the given phenotype. **P<0.001 (Fisher's exact test). (D,E) Whole-mount IHC for spi1b:EGFP in WT (n=11) and spag6 mutants (n=8). (F,G) Whole-mount ISH for the neutrophil marker mpx in WT (n=45) and spag6 mutant (n=11) embryos at 24 hpf. (H,I) Whole-mount ISH for the macrophage marker mfap4 in WT (n=19) and spag6 mutant (n=8) embryos at 24 hpf. (J-L) Quantification of myeloid progenitors (spi1b:EGFP+), neutrophils (mpx+) and macrophages (mfap4+) from a single yolk hemisphere of the individual embryos. (M,N) Representative confocal projection images from wounding experiments of WT sibling (n=4) and spag6−/− (n=4) mpx:EGFP transgenic embryos at 24 hpf. (O) Migration efficiency scores from cell tracks of yolk wounding assays in WT and spag6 mutant mpx:EGFP+ embryos at 24 hpf. ***P<0.0001, *P<0.05 (two-tailed unpaired Student's t-test). Scale bars: 50 μm (B-Ciii), 300 μm (F-I), 100 μm (D,E,M,N). PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|