- Title
-
Integrated Metabolomics and Lipidomics Analysis Reveal Remodeling of Lipid Metabolism and Amino Acid Metabolism in Glucagon Receptor-Deficient Zebrafish
- Authors
- Bai, X., Jia, J., Kang, Q., Fu, Y., Zhou, Y., Zhong, Y., Zhang, C., Li, M.
- Source
- Full text @ Front Cell Dev Biol
|
Profile of global metabolomics and lipidomics data. |
|
Pathway analysis of different metabolites. |
|
GCGR knockout induced glycerophospholipid metabolism dysregulation. |
|
GCGR knockout influenced arachidonic acid metabolism. Disturbed arachidonic acid metabolism. Dots represent metabolites, and blocks represent transcripts-encode enzymes. Red: up-regulated, blue: down-regulated; or green: set substrates mixed with both up-regulated and down-regulated metabolites. |
|
GCGR knockout influenced cholesterol metabolism. Dots represent metabolites, and blocks represent transcripts-encode enzymes. Red: up-regulated, blue: down-regulated; or green: set substrates mixed with both up-regulated and down-regulated metabolites. The heatmap indicates altered bile acids. Z score normalized ionic strength for each metabolite was represented by different colors: high (red), low (blue), or average (white). |
|
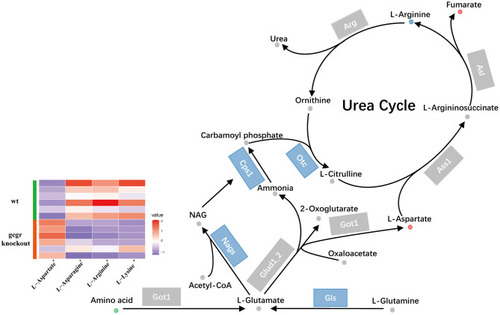
GCGR knockout influenced ureagenesis. Dots represent metabolites, and blocks represent transcripts-encode enzymes. Red: up-regulated, blue: down-regulated; or green: set substrates mixed with both up-regulated and down-regulated metabolites. The heatmap indicates altered amino acids. |
|
GCGR knockout influenced the tryptophan metabolism and locomotor activity in zebrafish. |
|
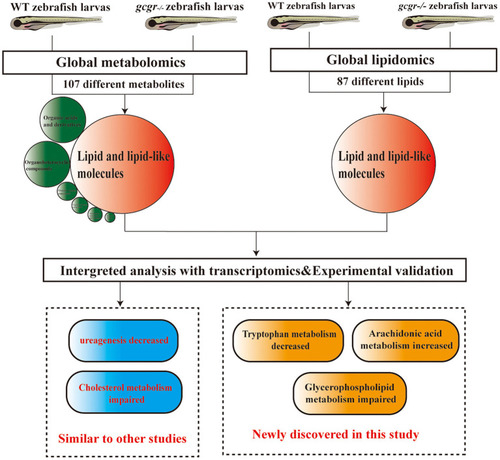
Summary of the experimental workflow and findings. Global metabolomics and lipidomics analysis were performed to study the metabolic change of |