- Title
-
Yulink, predicted from evolutionary analysis, is involved in cardiac function
- Authors
- Kuo, M.W., Tsai, H.H., Wang, S.H., Chen, Y.Y., Yu, A.L., Yu, J.
- Source
- Full text @ J. Biomed. Sci.
|
Structural features of Yulink protein. |
|
Characterization of EXPRESSION / LABELING:
PHENOTYPE:
|
|
|
|
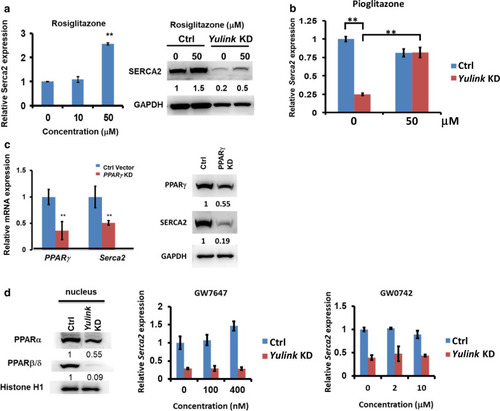
Over-expression of |
|
Effect of |
|
Yulink regulated |
|
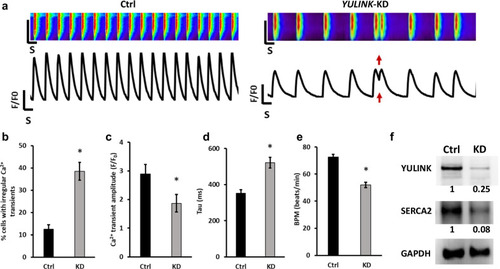
Assessment of arrhythmia and irregular Ca2+ regulation in |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|