- Title
-
Three-dimensional bright-field microscopy with isotropic resolution based on multi-view acquisition and image fusion reconstruction
- Authors
- Calisesi, G., Candeo, A., Farina, A., D'Andrea, C., Magni, V., Valentini, G., Pistocchi, A., Costa, A., Bassi, A.
- Source
- Full text @ Sci. Rep.
|
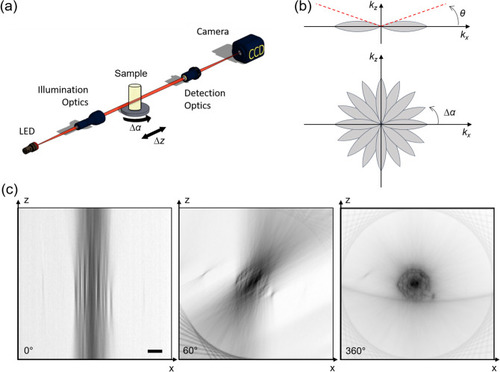
Experimental setup for multi-view bright-field reconstruction: an LED (530 nm) illuminates the sample, the transmitted light is collected by a detection optical system and a camera. The sample is mounted on a translation and a rotation stage to be scanned and rotated around 360°. A stack of images is acquired in each angular position while scanning the sample ( |
|
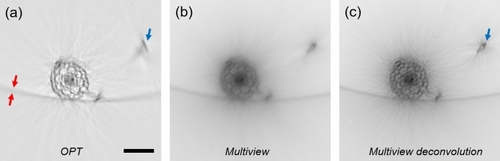
Reconstruction of a transverse section of an |
|
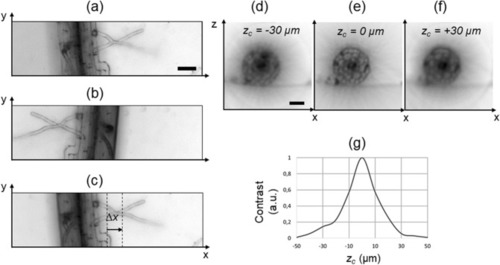
Search of the rotation axis. Minimum intensity projection (each pixel shows the minimum value of the stack calculated along the z direction) of the stack acquired at the angle 0° ( |
|
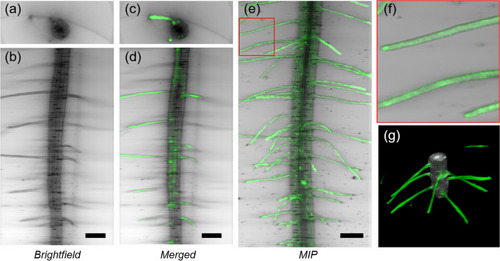
Three-dimensional imaging of a transgenic pEXP7:YC3.6 |
|
Sagittal slice of a transgenic Tg(kdrl:GFP) zebrafish (4 dpf) visualized with bright-field multi-view reconstruction ( |