- Title
-
Zebrafish (Danio rerio) physiological and behavioural responses to insect-based diets: a multidisciplinary approach
- Authors
- Zarantoniello, M., Randazzo, B., Gioacchini, G., Truzzi, C., Giorgini, E., Riolo, P., Gioia, G., Bertolucci, C., Osimani, A., Cardinaletti, G., Lucon-Xiccato, T., Milanović, V., Annibaldi, A., Tulli, F., Notarstefano, V., Ruschioni, S., Clementi, F., Olivotto, I.
- Source
- Full text @ Sci. Rep.
|
Zebrafish fed experimental diets specific growth rate (% weight growth day−1). Zebrafish fed diets including 0, 25, 50, 75 and 100% of BSF meal (Control, Hi25, Hi50, Hi75 and Hi100, respectively). Boxplots show minimum and maximum (whiskers), first quartile, median and third quartile (box). Different letters indicate statistically significant differences among experimental groups ( |
|
Content of SFA, MUFA and PUFA fatty acid (as % of total FA) and omega 3 (n3), omega 6 (n6) and omega 9 (n9) fatty acid contribution to lipid profile. ( |
|
Zebrafish (Control, Hi25,50,75,100 groups) intestine and liver histology and percentage of fat fraction (PFF) in liver tissue. |
|
( |
|
( |
|
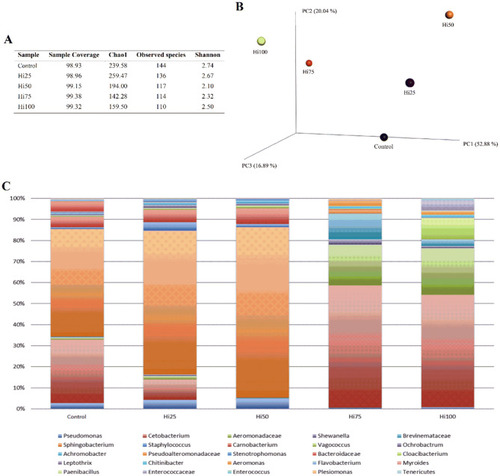
Microbiome of zebrafish fed diets including 0, 25, 50, 75 and 100% of BSF meal (Control, Hi25, Hi50, Hi75 and Hi100, respectively). ( |
|
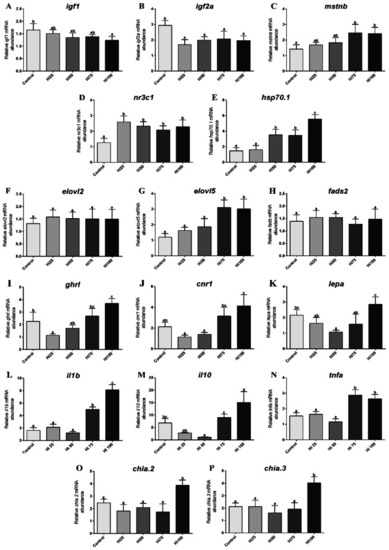
Relative mRNA abundance of genes analyzed in zebrafish fed on the different experimental diets (Control, Hi25, Hi50, Hi75 and Hi100). ( |
|
Behaviour |