Fig. 4
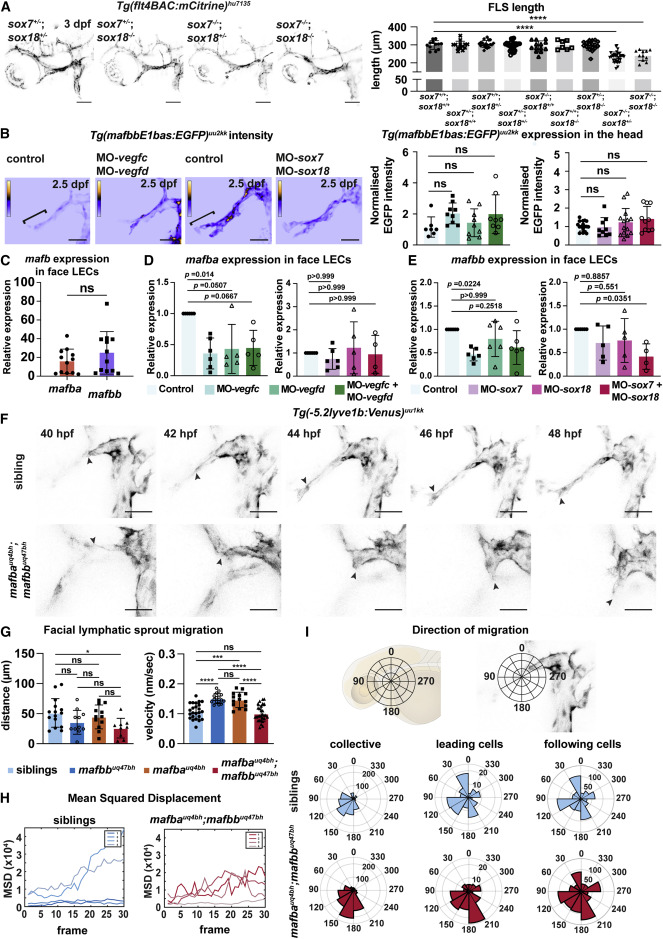
Figure 4. mafba and mafbb regulate facial LEC migration downstream of Vegfc-Vegfd-SoxF (A) (Left) Confocal images of Tg(flt4BAC:mCitrine) expression in facial lymphatics in sibling, sox7+/−;sox18−/−, sox7−/−;sox18+/−, and sox7−/−;sox18−/− embryos at 3 dpf. ∗: reduced sprout length. Scale bars, 100 μm. (Right) Quantification of facial lymphatic sprout length at 3 dpf in sox7 and sox18 mutant backgrounds. sox7+/+;sox18+/+ n = 12; sox7+/−;sox18+/+ and sox7−/−;sox18+/+ n = 13; sox7+/+;sox18+/− n = 18; sox7+/−;sox18+/− n = 30; sox7+/+;sox18−/− n = 7; sox7+/−;sox18−/− n = 17; sox7−/−;sox18+/− n = 32; sox7−/−;sox18−/− n = 11. One-way ANOVA: ∗∗∗∗p < 0.0001 for sox7+/+;sox18+/+ versus sox7−/−;sox18+/− or sox7−/−;sox18−/−; other comparisons: p > 0.9782. (B) (Top) Heatmap of Tg(mafbbE1bas:EGFP) signal intensity from confocal projection at 2.5 dpf in face LECs. Gray value intensity scale: 0–113 for control and MO-vegfc + MO-vegfd, scale: 0–147 for control and MO-sox7+MO-sox18. White bracket: quantification area. Scale bar, 50 μm. (Bottom) Quantification of Tg(mafbbE1bas:EGFP) intensity. (Left) Control n = 7; MO-vegfd and MO-vegfc n = 9; MO-vegfc + vegfd n = 8; (right) control n = 17; MO-sox7 n = 9; MO-sox18 n = 14; MO-sox7 + sox18 n = 10. Kruskal-Wallis test: ns p > 0.2372. (C) Quantitative real-time PCR of mafba and mafbb expression relative to cdh5 in facial LECs at 48 hpf. 12 replicates. Wilcoxon test: ns p = 0.3013. (D and E) Quantitative real-time PCR of mafba and mafbb expression in face LECs at 48 hpf. (D, left) 6 replicates for control and MO-vegfc and 5 for MO-vegfd and MO-vegfc + vegfd; expression relative to cdh5. (D, right) 6 replicates for control, MO-sox7, 5 for MO-sox18, and 4 for MO-sox7+MO-sox18; expression relative to B actin. (E, left) 6 replicates for control, MO-vegfd, MO-vegfc, and MO-vegfc + vegfd; expression relative to cdh5. (E, right) 6 replicates for control, 5 for MO-sox7 and MO-sox18, and 4 for MO-sox7+MO-sox18; expression relative to B actin. Kruskal-Wallis: p values reported on the graph. (F) Confocal projections from time-lapse imaging of LEC migration in siblings and double mutants labeled by Tg(-5.2lyve1b:Venus) from 40–48 hpf. Arrows: leading cells in FLS. Scale bars, 50 μm. (G) (Left) FLS migratory distance (from F). Embryos: siblings n = 15, mafbb n = 11, mafba n = 12; double mutants n = 9. One-way ANOVA: ∗p = 0.0279 for siblings versus double mutants; ns for all other comparisons. (Right) FLS migratory velocity (from F). Cells: siblings n = 24, mafbb n = 16, mafba n = 14; double mutants n = 29. One-way ANOVA: ∗∗∗∗p < 0.0001 for siblings versus mafbb, and mafbb or mafba versus double mutants; ∗∗∗p = 0.0001 for siblings versus mafba; ns for all other comparisons. (H) FLS tip mean-squared displacement (MSD) (from F) in siblings (n = 4) and double mutants (n = 4). (I) (Top) Schematic representation of directionality in the polar histograms. (Bottom) Polar histograms of migration tracked (from F) in siblings (n = 4) and double mutants (n = 4). Collective cells: siblings n = 24 and double mutants n = 29; leading cells: siblings n = 4 and double mutants n = 4; following cells: siblings n = 20 and double mutants n = 25. ns, non-significant; stars are significant; mean with SD.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.