Fig. 4
- ID
- ZDB-FIG-220628-157
- Publication
- Bortolin-Cavaillé et al., 2022 - Probing small ribosomal subunit RNA helix 45 acetylation across eukaryotic evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
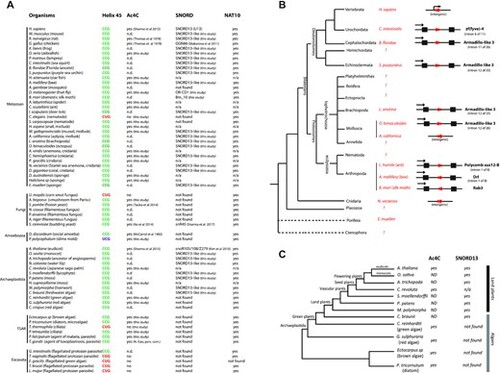
Cross-evolutionary survey of eukaryotic rRNA acetylation machinery. (A) This table summarizes the conservation of the 5′-CCG-3′ motif, the acetylation status of helix 45 and the detection of SNORD13 and NAT10 genes in commonly used models or organisms for which we experimentally assayed the presence of ac4C at helix 45. n.d.: not determined. n/a: not applicable (genome is not available) (56,57). (B) Simplified phylogenetic tree of Metazoan. The genomic organization (intronic vs intergenic) of newly-identified SNORD13 genes (red arrows) is shown. Note that intronic SNORD13 can be positioned in the sense or antisense orientation with respect to their host-genes (black arrow). (C) Simplified phylogenetic tree of Archaeplastida. The acetylation status of helix 45 and the presence of SNORD13 in the genome of representative species are indicated. Note that while rRNA in algal species is acetylated, we failed to identify obvious SNORD13 counterparts. A full list of newly-identified SNORD13 sequences can be found in Supplementary data S1. |