Fig. 1
- ID
- ZDB-FIG-220628-154
- Publication
- Bortolin-Cavaillé et al., 2022 - Probing small ribosomal subunit RNA helix 45 acetylation across eukaryotic evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
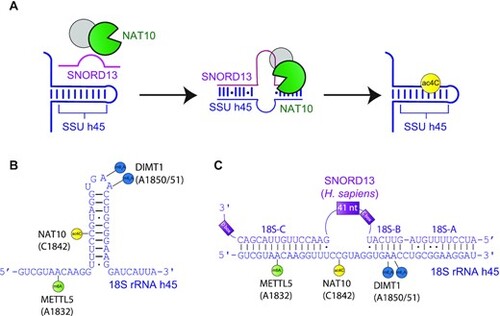
Schematic of SNORD13-dependent ribosomal RNA acetylation. (A) Through an incompletely defined antisense-mediated mechanism, human SNORD13 assists the RNA cytidine acetyltransferase NAT10 in modifying C1842 in the universally conserved helix 45 at the 3′ extremity of 18S rRNA. (B) Helix 45 is enriched with post-transcriptional RNA modifications. RNA modifying enzymes and their modified nucleotides (lollipops) are indicated: SSU-ac4C1842 (yellow), SSU-m6A1832 (green) and SSU-m26A1850 and SSU-m26A1851 (blue) are introduced by NAT10, METTL5 and DIMT1, respectively. (C) Human SNORD13 base-pairs with 18S rRNA sequences on each side of the cytidine to be acetylated. Note that SNORD13-rRNA duplexes tolerate G-U wobble base-pairs, mismatches or even bulged nucleotides and the length of the intervening loops, from either rRNA or SNORD side, also differ from one organism to another (Supplementary Figure S1). |