Fig 3
- ID
- ZDB-FIG-211207-25
- Publication
- Marsay et al., 2021 - Tetraspanin Cd9b and Cxcl12a/Cxcr4b have a synergistic effect on the control of collective cell migration
- Other Figures
- All Figure Page
- Back to All Figure Page
|
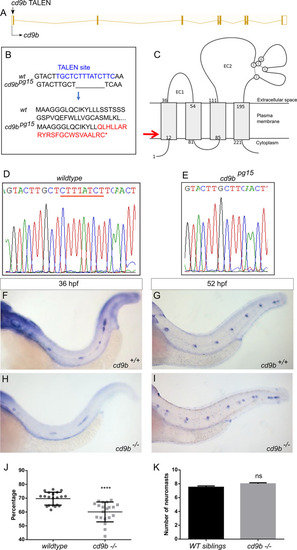
cd9b mutant does not recapitulate morphant phenotype, although primordium migration is delayed at 36 hpf.
A: Nature of the cd9b mutant allele showing TALEN site location within the intron-exon structure of the gene. B: The TALEN target sequence in exon 1 is shown in blue; the 8bp deletion in the cd9b allele is indicated under the WT sequence as dashes. This leads to a frameshift changing codon 15 from TTT (Phe) to CAA (Glu), then 22 aberrant amino acids (red lettering) followed by a stop codon (*). C: Schematic of the Cd9b protein with location of premature stop codon given by red arrow. The disulfide bonds between the conserved CCG motif and conserved cysteines are indicated by the dashed lines. EC1/2 = Extracellular domain 1/2, aa = amino acid. D-E: Sequence chromatograms of genomic DNA from (d) WT and (e) cd9bpg15 alleles. Deleted base pairs are underlined in red. F-I: Representative images of WISH of claudin b ISH on (f-g) WT and (h-i) cd9b mutants at time shown. J-K: Graphs quantifying pLLP measurements in WT and cd9b KOs; (j) the migration of the cd9b KO primordium at 36 hpf is significantly delayed compared to WT. (k) There is no significance in number of neuromasts deposited at 52 hpf. Unpaired T test on untransformed data. n = minimum 20. **** = p = <0.05. |