Fig. 1
- ID
- ZDB-FIG-211201-257
- Publication
- Winter et al., 2021 - Functional brain imaging in larval zebrafish for characterising the effects of seizurogenic compounds acting via a range of pharmacological mechanisms
- Other Figures
- All Figure Page
- Back to All Figure Page
|
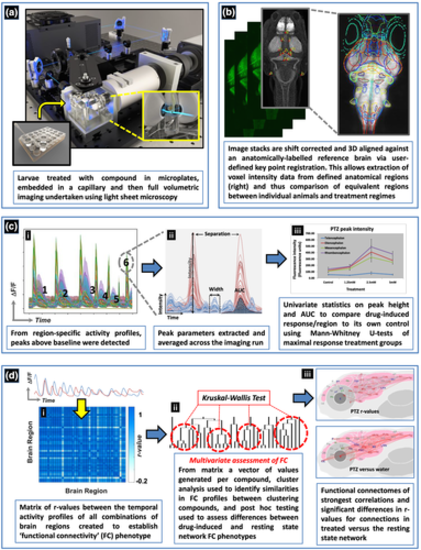
Schematic of the experimental process used to obtain 4D whole brain neural activity data from drug treated larval elavl3:GCaMP6s zebrafish. The experimental process and resultant data analysis procedures are summarised in order of panels a–d. (a) Animals were first exposed to the compound and then imaged using light sheet microscopy. (b) The resultant image stacks were 3D aligned to a standardised brain image library (Z-brain atlas) and subsequent 3D registration allowed extraction of anatomical region-specific fluorescence data. (c) The extracted region-specific fluorescence intensity temporal profiles were then analysed to reveal regional changes between treated and untreated animals. (i) Peaks were automatically identified from the temporal profiles (e.g. labelled 1–6) and (ii) peak parameters for each profile extracted. (iii) Peak height and area under the curve (AUC) were then analysed using univariate statistics to identify brain regions showing altered neural activity versus the same area in the corresponding group of control animals. (d) For the analysis of functional connectivity (FC), (i) Pearson's correlation coefficients (r) were calculated between the temporal activity profiles of each pair of regions to provide a matrix summarising measures of the level of functional connectivity between each pair of nodes. An r value of >0.7 between two regions suggested these areas were functionally connected as both showed correlated temporal activity profiles. (ii) Multivariate statistical analysis of the FC data was then undertaken, followed by cluster analysis of the resultant Euclidean distances, to assess similarities in the resultant phenotypes and whether groupings contained compounds sharing common mechanisms of pharmacological action. Post hoc testing of the resultant groupings using Kruskal–Wallis tests then revealed the brain regions and/or connections driving these similarities/differences. (iii) The r values from the FC matrices generated for each compound were visualised in the form of a connectivity map of the brain (a functional connectome ~ upper panel) and as exposed animal data versus the resting state network by comparing the r values for each connection between treated and untreated groups using Mann–Whitney U tests (lower panel) |