Fig. 1
- ID
- ZDB-FIG-210923-18
- Publication
- Neal et al., 2021 - ETS factors are required but not sufficient for specific patterns of enhancer activity in different endothelial subtypes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
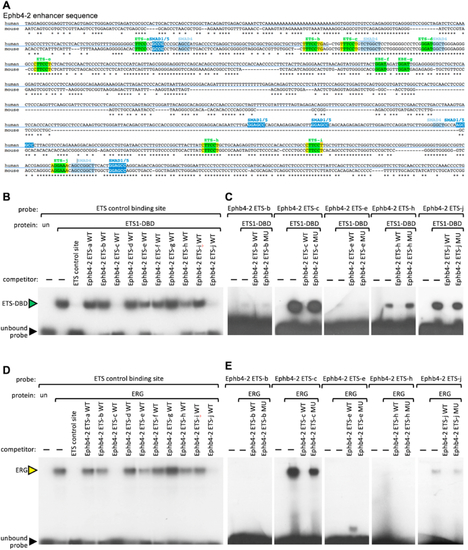
The Ephb4-2 venous enhancer contains functional ETS motifs. A. ClustalW alignment of the human and mouse sequences of the Ephb4-2 enhancer annotated with conserved ETS binding motifs (green), SMAD4 binding motifs (light blue) and SMAD1/5 binding motifs (dark blue) as previously reported ( |
Reprinted from Developmental Biology, 473, Neal, A., Nornes, S., Louphrasitthiphol, P., Sacilotto, N., Preston, M.D., Fleisinger, L., Payne, S., De Val, S., ETS factors are required but not sufficient for specific patterns of enhancer activity in different endothelial subtypes, 1-14, Copyright (2021) with permission from Elsevier. Full text @ Dev. Biol.