|
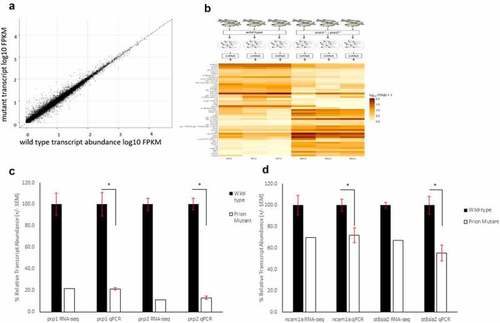
RNA-Sequencing show 1249 genes with an increase or decrease of log2 fold change of 0.5 between wild-type and compound homozygous prion mutant zebrafish larvae. (A) Scatter graph showing relative FPKM values for wild-type (X-axis) and mutant (Y-axis) genes after RNA-sequencing. (B) Methodology diagram showing RNA-sequencing workflow. Two groups, wild-type and prion mutant (prp1ua5003/ua5003; prp2ua5001/ua5001) homozygous fish, each with three replicates containing a pool of 50 3dpf larvae were processed and sent for RNA-sequencing. Heatmap displays sample of the 25 genes with a biggest differential abundance in FPKM in wild-types compared to mutants. (C) Relative transcript abundance between wild-type and prion mutant fish comparing prp1 and prp2 through both RNA-sequencing and RT-qPCR analysis. D) Relative transcript abundance of ncam1a and st8sia2 through both RNA-sequencing and RT-qPCR analysis. * = P < 0.05
|