FIGURE 1
- ID
- ZDB-FIG-210518-12
- Publication
- Cheng et al., 2021 - Unbiased Label-Free Quantitative Proteomics of Cells Expressing Amyotrophic Lateral Sclerosis (ALS) Mutations in CCNF Reveals Activation of the Apoptosis Pathway: A Workflow to Screen Pathogenic Gene Mutations
- Other Figures
- All Figure Page
- Back to All Figure Page
|
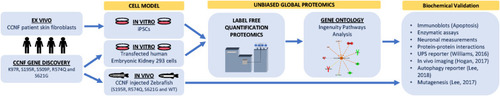
Proteogenomic workflow to determine whether potential ALS gene candidates cause disease. Discovery of gene mutations by whole-genome, whole-exome, or targeted sequencing. cDNA gene constructs candidate mutations are inserted into cell models (e.g., HEK293 or Neuro2A cells). Ex vivo models such as patient fibroblasts (if available) that contain the gene mutation can also be used or converted into iPSCs for further differentiation. Label-free quantitative proteomics is used to analyze and provide a profile of protein expression changes that cluster to biological processes and signalling pathways. These biological pathways can then be validated using standard biochemical methods and/or in animal models. Animal models, such as zebrafish, have the advantage of generating progeny relatively quickly and can be extended to study motor phenotypes. |