Figure 1
- ID
- ZDB-FIG-200314-68
- Publication
- Kang et al., 2020 - Global Transcriptomic Analysis of Zebrafish Glucagon Receptor Mutant Reveals Its Regulated Metabolic Network
- Other Figures
- All Figure Page
- Back to All Figure Page
|
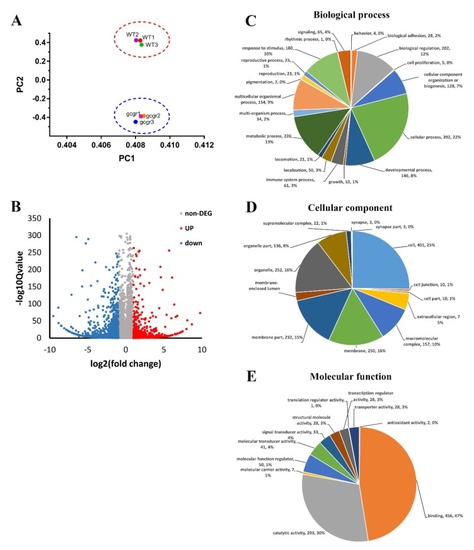
RNA-seq (RNA sequencing) analysis of glucagon receptor ( |