|
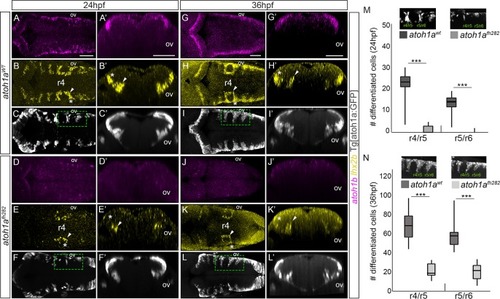
<italic>atoh1a</italic> is required for the specification of the <italic>lhx2b</italic> neuronal population.A-L) atoh1aWT and atoh1afh282 embryos in the Tg[atoh1a:GFP] background were analyzed at 24hpf (atoh1aWT n = 14; atoh1afh282 n = 18) and 36hpf (atoh1aWT n = 11; atoh1afh282 n = 7) with atoh1b (A, D, G, J), lhx2b (B, E, H, K), and anti-GFP in order to follow the atoh1a-derivatives (C, F, I, L). A’-L’) Reconstructed transverse views of dorsal views displayed in (A-L) at the level of the anterior side of the otic vesicle. Note that atoh1b expression (compare A-A’ and G-G’ with D-D’ and J-J’), and the lateral domains of lhx2b diminished (compare white asterisks in B-B’ with E-E’, and H-H’ with K-K’), whereas the more medial domain does not decrease so dramatically (compare white arrowheads in B-B’ with E-E’, and H-H’ with K-K’). Note that atoh1a:GFP cells remained, suggesting that there is no massive cell death. M-N) Quantification of differentiated neurons in the r4/r5 and r5/r6 domains of atoh1aWT and atoh1afh282 embryos as depicted in the small inserts showing dorsal views of halves hindbrains that correspond to the framed regions in (F-L), *** p<0.001 (Table 1 for values and statistical analysis). Note the reduction in the number of atoh1a:GFP differentiated neurons in atoh1afh282 embryos. ov, otic vesicle; r, rhombomere. Scale bars correspond to 50 μm.
|