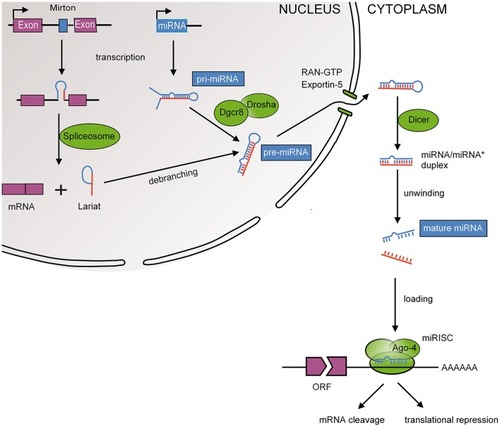
MiRNA biogenesis. Mature miRNAs are encoded in the genome and form after a series of enzymatic cleavages from two possible precursor molecules; primary miRNAs (pri-miRNAs) or Mirtrons. Pri-miRNAs, following the canonical pathway, are transcribed as long hairpin RNAs that are recognized by the RNA-binding DiGeorge syndrome critical region 8 protein (DGCR8). Many pri-miRNAs are often transcribed simultaneously due to clustering of several miRNA genes (Ambros et al., 2003). DGCR8 then directs the RNase III endonuclease DROSHA to cleave the base of the hairpin to produce ~70 nucleotide hairpins known as pre-miRNA. Mirtrons, following the non-canonical pathway, bypass the microprocessor as they are transcribed as part of the introns of protein coding genes and are as such spliced by the spliceosome (Berezikov et al., 2007). Splicing also produces ~70 nucleotide hairpins known as pre-miRNA. The pre-miRNA is transported to the cytoplasm by exportin 5 where it is cleaved by another RNase III endonuclease known as DICER to ~20 nucleotide miRNA duplexes with protruding 2 nucleotide 3′ ends. The resulting mature miRNA is released and a guiding strand is incorporated into the RNA-induced silencing complex (RISC).
|