Fig. 4
- ID
- ZDB-FIG-180620-75
- Publication
- Murphy et al., 2018 - Placeholder Nucleosomes Underlie Germline-to-Embryo DNA Methylation Reprogramming
- Other Figures
- All Figure Page
- Back to All Figure Page
|
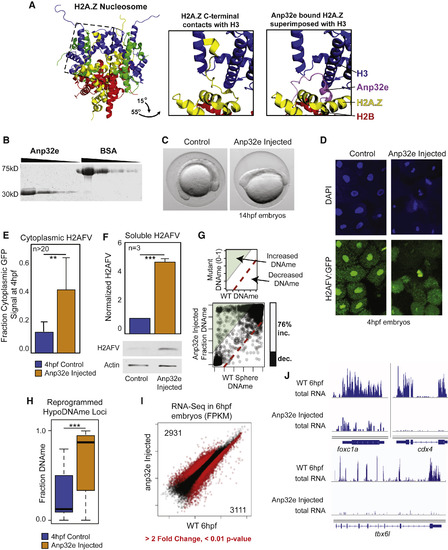
Anp32e Protein Injection Causes Reduced Nuclear H2AFV and Increased DNA Methylation (A) Anp32e binding inhibits H2A.Z within the nucleosome. Left: crystal structure for the nucleosome octamer containing mouse H2A.Z (yellow) (Suto et al., 2000). Middle: zoomed-in and rotated view of the H2A.Z nucleosome octamer (dashed box from left), where the C terminus of H2A.Z interacts with histone H3. Right: the same view as middle panel, with the superimposed H2A.Z bound to human ANP32E (Obri et al., 2014). (B) Purification of recombinant zebrafish Anp32e protein. A comparison to BSA and Coomassie staining to demonstrate purity. (C) Phenotype of early somite-staged zebrafish following Anp32e protein injection. (D) H2AFV signal is depleted in nuclei following Anp32e injection in embryos. DAPI and H2AFV:GFP localization in cells from sphere-staged (4hpf) embryos. Injection was into 1-cell embryos. (E) Microscopy-based quantification of H2AFV following Anp32e injection. Fraction H2AFV:GFP signal in control and injected embryos like in (D). t tests: ∗∗ p < 0.01 (F) Quantification of H2AFV in cellular fractions resulting from Anp32e injection. Western blotting for soluble H2AFV indicates that Anp32e injection causes increased cytoplasmic H2AFV. t tests: ∗∗∗ p < 0.001. (E and F) Error bars represent SD. (G) Anp32e injection causes increased DNAme. Scatterplot of CpGs for treatment versus control is displayed, with a bar graph quantifying changing CpGs (>20% absolute change). (H) Changes in DNA methylation are most significant at regions at which DNA is normally demethylated. t tests: ∗∗∗ p < 0.001. Boxplots are displayed. (I) A scatterplot of gene transcript abundance (fragments per kilobase of million mapped reads [FPKM]) for Anp32e-injected embryos compared to control samples. Red data points indicate fold change >2 and p < 0.01 false discovery rate (FDR). The number of genes changing is indicated in the top-left and bottom-right corners. (J) A gallery of browser snapshots for total RNA at developmentally regulated genes. See also Figure S4. |
Reprinted from Cell, 172(5), Murphy, P.J., Wu, S.F., James, C.R., Wike, C.L., Cairns, B.R., Placeholder Nucleosomes Underlie Germline-to-Embryo DNA Methylation Reprogramming, 993-1006.e13, Copyright (2018) with permission from Elsevier. Full text @ Cell