Fig. S1
- ID
- ZDB-FIG-160324-22
- Publication
- Hoffman et al., 2016 - Estrogens Suppress a Behavioral Phenotype in Zebrafish Mutants of the Autism Risk Gene, CNTNAP2
- Other Figures
- All Figure Page
- Back to All Figure Page
|
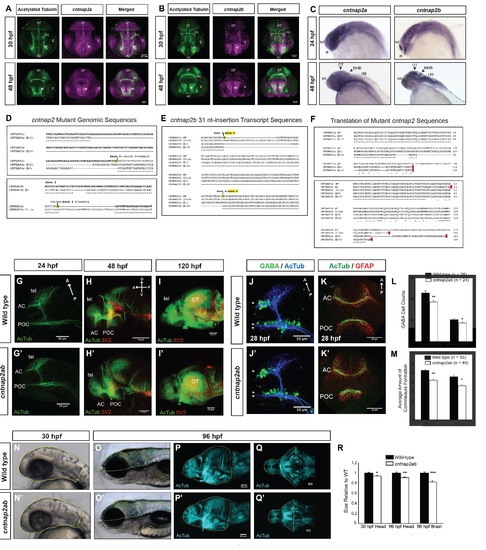
Figure S1, Related to Figure 1. Generation of cntnap2ab mutants and characterization of brain structure. A, B. Fluorescent in situ hybridization of cntnap2a and cntnap2b expression and acetylated tubulin in wild-type embryos at 30 and 48 hpf. C. Whole mount in situ hybridization of cntnap2a and cntnap2b in wild-type embryos at 24 and 48 hpf. Both paralogs are expressed in the CNS, though cntnap2b shows a more specific expression pattern in the telencephalon and diencephalon (24 hpf), the optic tectum, midbrain-hindbrain boundary, and hindbrain (48 hpf). D. Genomic sequences of insertion-deletion mutations generated by zinc finger nucleases in each cntnap2 paralog. These include Δ121 and Δ25 in exon 3 of cntnap2a (top) and Δ7 in exon 2 and a 31-nt insertion in exon 4 of cntnap2b (bottom), all of which produce a frameshift and premature stop codon. E. Alternatively spliced transcripts resulting from the 31-nt allele in exon 4 of cntnap2b identified by RT-PCR of exons 1 to 6 at 5 dpf. These transcripts include: (i) the 31-nt sequence itself, as in the genomic sequence; (ii) Δ10 in exon 4: because the 31-nt insertion occurs close to the intron-exon 4 boundary, an alternate splice site is used (the next “TCAG” in the genomic sequence after the actual intronic splice site), which is 10 nt after the beginning of exon 4; (iii) Δ148 due to splicing of the entire exon 4. All 3 transcripts produce a frameshift and a premature stop codon. We did not identify alternatively spliced transcripts resulting from the other mutations in either gene. F. Sequence alignment of wild type cntnap2a (top) and cntnap2b (bottom) proteins and the translation of mutant transcripts. All mutant transcripts are predicted to cause premature stop codons in the discoidin domains of both proteins (Figure 1A). G-I. Acetylated tubulin (AcTub) and synaptic vesicle protein 2 (SV2) labeling in wild-type (G-I) and cntnap2aΔ121/Δ121cntnap2b31i/31i (G′-I′) fish at 24, 48, and 120 hpf. While there is a transient delay in the formation of the anterior and post-optic commissures in cntnap2ab mutants at 28 hpf (see K, K′, M), these commissures form by later stages. Brain structure is not otherwise grossly disrupted in cntnap2ab mutants up to 120 hpf. J-K. Acetylated tubulin and GABA (J, J′) or GFAP (glial cells) (K, K′) in wild-type and cntnap2aΔ121/Δ121cntnap2b31i/31i (cntnap2ab) mutants at 28 hpf. L. There are significantly fewer GABA+ cells in both the telencephalon (arrowheads) and diencephalon (double arrowheads) of cntnap2ab (J′) versus wild-type (J) larvae at 28 hpf. GABA+ cells were counted in z-stack images by three blinded raters. (*p=0.00782, telencephalon; **p=0.0181, diencephalon, two-way ANOVA, genotype factor). M. The formation of the anterior commissure (AC) and post-optic commissure (POC) is delayed in cntnap2aΔ121/Δ121cntnap2b31i/31i (cntnap2ab) (K′) compared to wild-type (K) larvae (*p=0.00428, AC; **p=0.0159, POC, two-way ANOVA, genotype factor.) Three blinded raters quantified the degree of commissure formation based on a qualitative rating scale ranging from no axons crossing (1) to complete formation (7), as previously described (Barresi et al., 2005). The POC appears to be less fasciculated in mutants. Glial bridge formation is largely intact. N-R Head and brain size in wild-type (N-Q) and cntnap2aΔ121/Δ121cntnap2b31i/31i (cntnap2ab) (N′-Q′). Representative tracings done in Fiji are shown in yellow. The posterior boundaries of the head and brain were defined by the otic vesicle and the midbrain-hindbrain boundary, respectively. R. cntnap2ab mutant heads are 6.5% and 9.7% smaller than wild-type at 30 hpf and 96 hpf, respectively, and mutant brains are 18% smaller than wild-type at 96 hpf. (*p=9.71x10-7, 30 hpf head size; **p=0.00513, 96 hpf head size; ***p=3.89x10-6, 96 hpf brain size; one-way ANOVA). In addition, eye and yolk size measurements indicate that mutant fish are smaller overall (Table S1). (A, B): frontal views; (C, N-O, N′-O′): lateral views; (G-J, G′-J′, P-P′): lateral views of dissected embryos with the eye removed; (K-K′): frontal views of the forebrain; (Q-Q′): ventral views; tel, telencephalon; di, diencephalon; OT, optic tectum; PC, posterior commissure; AC, anterior commissure; POC, post-optic commissure; E, eye; MHB, midbrain-hindbrain boundary; HB, hindbrain. |