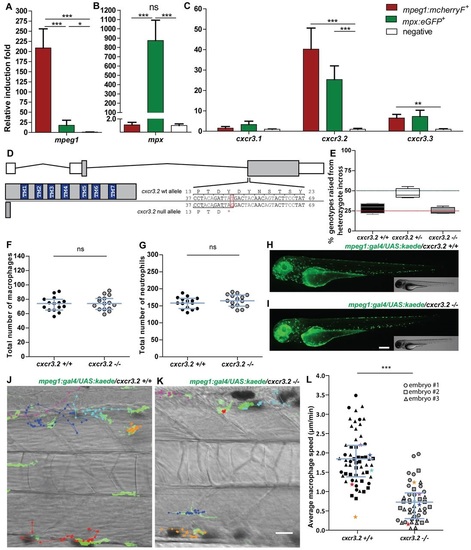
Characterization of cxcr3.2−/− embryos in unchallenged conditions. (A–C) Expression of cxcr3.2 and its paralogs cxcr3.1 and cxcr3.3 in FACS-sorted phagocytes. Graphs represent the relative induction fold of the macrophage marker mpeg1 (A), the neutrophil marker mpx (B), and of the cxcr3 paralogs (C) in FACS-sorted macrophages and neutrophils from the combined transgenic line Tg(mpeg1:mcherryF/mpx:eGFP) at 2 dpf. Expression of cxcr3.2 and cxcr3.3 could be detected in both macrophages and neutrophils, whereas cxcr3.1 was not significantly enriched in the FACS-sorted populations when compared with the non-labeled cell fraction. Sample size (n): five replicates. Errors bars: mean±s.e.m. Reference gene: eif4a1b. (D) Effect of the cxcr3.2 point mutation. Top: gene structure of cxcr3.2. Boxes represent exons, of which the gray parts correspond to the coding sequence. Bottom right: cxcr3.2 wild-type (wt) and mutant allele. A single T-to-G mutation at nucleotide 48 generates an early stop codon. Bottom left: consequence of cxcr3.2 mutation at the protein level. In cxcr3.2 mutant zebrafish, only a peptide of 15 amino acids can be translated, which lacks all the conserved transmembrane domains (TM1-7). Nucleotide and amino acid positions are enumerated from the translation start codon. (E) Normal viability of cxcr3.2−/− mutants. Percentage of genotypes deriving from cxcr3.2+/− incross. No significant deviation from the Mendelian 1:2:1 ratio was observed. The genotypes were evaluated on 122 adult fish from four independent breedings. The boxplots represent the area of the distributions between the first and the third quartiles. Whiskers represent the minimum and maximum end points of the distributions. (F,G) Quantification of macrophages and neutrophils. Combined Leukocyte-plastin (Lp) immunostaining and Myeloperoxidase (Mpx) staining were performed on cxcr3.2+/+ and cxcr3.2−/− embryos at 3 dpf and the numbers of stained cells residing in the caudal hematopoietic tissue were counted. Exclusively Lp-stained cells were considered as macrophages (F) and Lp/Mpx double-positive cells as neutrophils (G). No significant differences were detected. Total number of larvae (n) per group in both F and G: 15. Error bars: median and interquartile range. (H,I) Spatial distribution of macrophages. A macrophage-specific transgenic reporter driven by the mpeg1 promoter [Tg(mpeg1:gal4/UAS:kaede) (Ellett et al., 2011)] was crossed into the cxcr3.2 mutant background. Representative images of the resulting Tg(mpeg1:gal4/UAS:kaede) cxcr3.2+/+ (H) and cxcr3.2−/− (I) larvae at 3 dpf show no major differences in the macrophage distribution pattern. Scale bar: 200 μm. (J,K) Macrophage basal migratory capability. Paths of five representative macrophages in the trunk of Tg(mpeg1:gal4/UAS:kaede) cxcr3.2+/+ (J) and cxcr3.2−/− (K) larvae at 3 dpf. Mutant and wt larvae were mounted in agarose on the same dish and behavior of mutant and wt macrophages were simultaneously followed for 3 hours. Time-lapse images were taken every 6 minutes. The paths were followed and analyzed using ImageJ ManualTrack plugin. See also supplementary material Movies 1, 2. Scale bar: 20 μm. (L) Quantification of basal migration difference. The average speed of individual macrophages was calculated by tracking 15–21 macrophages from three different Tg(mpeg1:gal4/UAS:kaede) cxcr3.2+/+ and cxcr3.2−/− larvae (each larva is indicated with a different symbol) and was significantly reduced in cxcr3.2−/− macrophages. Total number of tracks (n): 61, 48. Error bars: mean and interquartile range. ns, non-significant; *P<0.05; ***P<0.001.
|

