Fig. 3
- ID
- ZDB-FIG-140813-7
- Publication
- Shimoda et al., 2005 - Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene
- Other Figures
- All Figure Page
- Back to All Figure Page
|
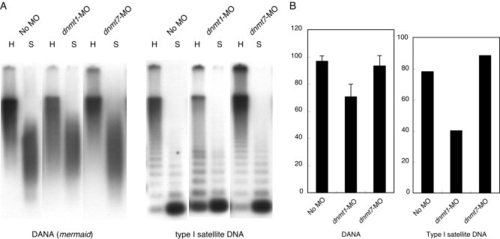
Methylation analysis of repetitive sequences. A: Southern blot analysis of methylation status of DANA/mermaid repeat and Type I satellite sequence. The enzyme, HpyCH4IV (H), which is present in both repeats, cannot cleave its recognition sequence of “ACGT” when its CpG is methylated. Compared with the control (No MO), increased cleavages of DANA and Type I with the enzyme were observed in the genomes of dnmt1-MO-injected embryos, reflecting substantial hypomethylation of CpGs. On the other hand, identical patterns were observed between the control and dnmt7-injected embryos, indicating no change in CpG methylation. Because isoschizomers of HpyCH4IV, which are not sensitive to CpG methylation, were not available, we used Sau3AI (S) to see how binding patterns were changed when methylation in these repeats was reduced, because Sau3AI digestion is less affected by CpG methylation than HpyCH4IV digestion. B: Quantitative analysis of methylation status of DANA/mermaid and Type I sequences. There are 15 CpG dinucleotides in the sequenced region in the DANA. Mean values and standard errors were determined based on 10 independent sequences. Because there is only one CpG site in Type I repeat, only mean values are shown for Type I. Template DNAs used for A and B were the same. |
| Fish: | |
|---|---|
| Knockdown Reagent: | |
| Observed In: | |
| Stage: | Long-pec |