- Title
-
Identification of a gene required for de novo DNA methylation of the zebrafish no tail gene
- Authors
- Shimoda, N., Yamakoshi, K., Miyake, A., and Takeda, H.
- Source
- Full text @ Dev. Dyn.
|
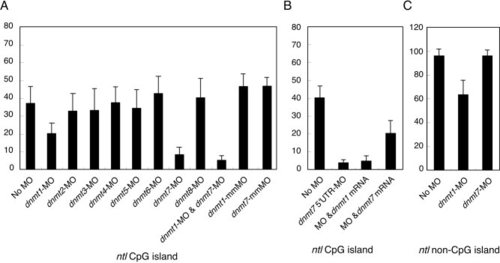
Suppression of Dnmt1 or Dnmt7 activity affects ntl methylation. A: Methylation status of the ntl CpG island was analyzed 48 hr after the injection of morpholino oligonucleotide (MO) against each dnmt. mm-dnmt1 and mm-dnmt7 are negative-control MOs carrying mismatch nucleotides in their original dnmt1-MO and dnmt7-MO sequences, respectively. Part of the ntl CpG island containing 16 CpG dinucleotides was PCR amplified and 10 independent clones were sequenced. Each column represents the mean ± standard error of methylated cytosines. B:ntl methylation level reduced by dnmt7 52UTR-MO was partially restored by coinjection of in vitro-synthesized dnmt7 mRNA (approximately 500 pg), which has no complementary sequence to the dnmt7 5′UTR-MO. Also, 200 pg of the same mRNA was unable to rescue the de novo methylation (data not shown). Because dnmt7 52 UTR-MO caused a neural death phenotype that at low doses is transient, we used less of the 52 UTR-MO (1.5 ng) than that of the original dnmt7-MO (2.0 ng). C: Methylation status of an ntl 32 exon region. An ntl 3′ region harboring 15 CpGs was amplified, and 10 independent clones were sequenced. PHENOTYPE:
|
|
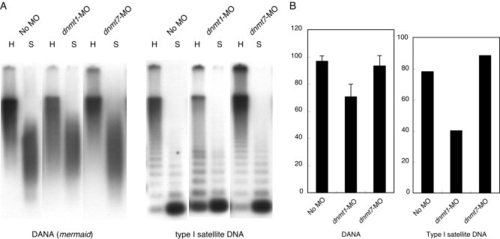
Methylation analysis of repetitive sequences. A: Southern blot analysis of methylation status of DANA/mermaid repeat and Type I satellite sequence. The enzyme, HpyCH4IV (H), which is present in both repeats, cannot cleave its recognition sequence of “ACGT” when its CpG is methylated. Compared with the control (No MO), increased cleavages of DANA and Type I with the enzyme were observed in the genomes of dnmt1-MO-injected embryos, reflecting substantial hypomethylation of CpGs. On the other hand, identical patterns were observed between the control and dnmt7-injected embryos, indicating no change in CpG methylation. Because isoschizomers of HpyCH4IV, which are not sensitive to CpG methylation, were not available, we used Sau3AI (S) to see how binding patterns were changed when methylation in these repeats was reduced, because Sau3AI digestion is less affected by CpG methylation than HpyCH4IV digestion. B: Quantitative analysis of methylation status of DANA/mermaid and Type I sequences. There are 15 CpG dinucleotides in the sequenced region in the DANA. Mean values and standard errors were determined based on 10 independent sequences. Because there is only one CpG site in Type I repeat, only mean values are shown for Type I. Template DNAs used for A and B were the same. PHENOTYPE:
|
|
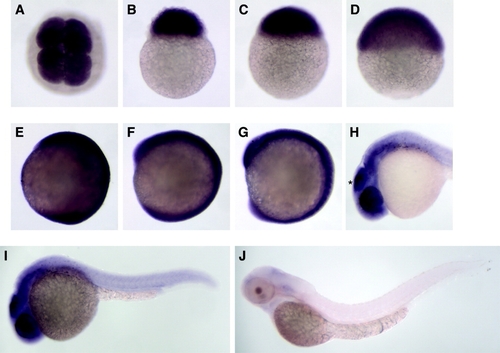
Expression pattern of dnmt7. A-J: The embryo stages used for in situ hybridization were 4 cells (A), 256 cells (B), 1,000 cells (C), 50% epiboly (D), tail bud (E), 5 somite (F), 10 somite (G), day 1 (H,I), and day 2 (J). A,B: dnmt7 mRNA could be detected before the mid-blastula stage (1,000 cells) of zebrafish embryos, indicating its maternal supply. F,G: During the somite stage, dnmt7 was ubiquitously expressed in zebrafish embryos, and its expression persisted until day 2. H,I: In 1-day-old embryos, higher expression was observed in the brain, especially in the tectum (asterisk in H). |
|
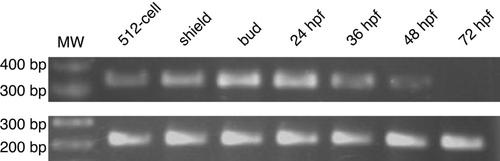
Reverse transcriptase-polymerase chain reaction (RT-PCR) of dnmt7 transcript. RNA derived from embryos at indicated stages was assayed by RT-PCR for the transcripts of dnmt7 (upper panel) and ubiquitously expressed EF1α (lower panel). MW, molecular weight marker. EXPRESSION / LABELING:
|