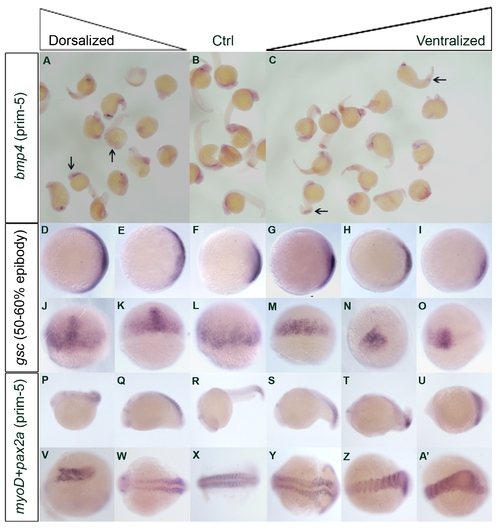
In situ hybridization results of selected genes at different stages of embryonic development after overexpression and knockdown experiment
Additional in situ hybridization experiments at different stages had been preformed to confirm the dorsalized or ventralized phenotypes after being examined by eyes. Panels A to C showed the bmp4 mRNA expression pattern at prim-5. Noted that there is a decrease of mRNA expression in the head region (arrows) for dorsalized embryos (A) when compared to control (B). Ventralized embryos (C) are featured with the ectopic expression at the tip of the yolk extension as well as the distal border of the expanded blood island (arrows). Panels D to I, animal pole view, dorsal towards the right; J to O, dorsal view, showed the gsc mRNA expression pattern at 50-60% epiboly. Different degree of expanded expression patterns were found in the dorsalized embryos (D, E, J, K). In contrast, restricted patterns were found in the ventralized embryos (G-I, M-O) when compared to control (F, L). Panels P to A′ showed the mRNA expression pattern of pax2a and myoD at prim-5 stage. R and X were the control. Noted that dorsalized embryos were characterized by shortened and uneven (P, V) and/or laterally expanded (Q, W) myoD expression. In ventralized embryos (S-U, Y-A′), myoD expression is stronger at the tail end. Abnormal tail development was indicated by fused myoD patterning. P to U, lateral views, anterior to the left; V to A′, dorsal view.
|

