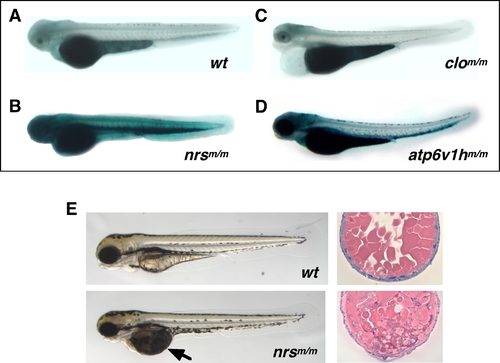
Mutant nrs zebrafish show extremely high SA-β-gal activity and display both yolk and muscle phenotypes.
3.5-day old (3.5 dpf) homozygous nrsm/m (hi891/hi891) zebrafish embryos show extremely high SA-β-gal activity (B) compared with wild-type embryos (A) (with PTU). The atp6v1hm/m (hi923/hi923) mutant shown in (D) is another variant identified as having significantly higher SA-β-gal activity. Most other early embryonic lethal mutants derived from either insertional mutagenesis (as shown in Figure S4) and chemical mutagenesis (e.g., clom39/m39 which is shown in (C)) show no higher (or indistinguishable) SA-β-gal activity than wild-type siblings at any time during development. (E) Yolk opaque phenotypes can be observed in homozygous nrsm/m embryos at 3.5 dpf (earliest detection at around 2.5 dpf; lower left panel), compared with wild-type embryos (upper left panel). Also shown is a comparison between the H&E staining of transverse sections of the yolk part of nrsm/m embryos at 3.5 dpf (lower right panel) and wild-type embryos (upper right).
|

