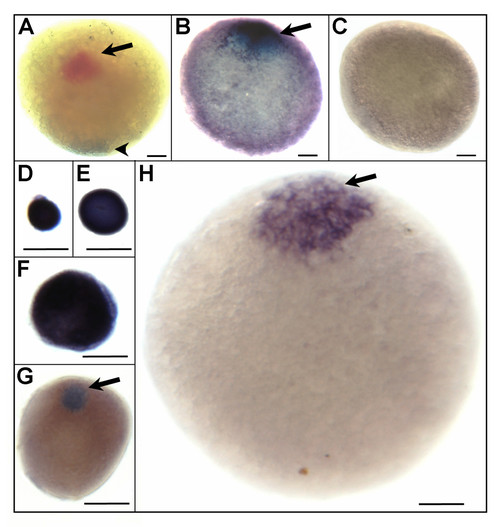
Localisation of selected maternal transcripts in zebrafish fully-grown follicles as revealed by whole-mount in situ hybridization. A, Polarization of the oocyte along the animal-vegetal axis was visualized using two colour whole-mount in situ hybridization. Cyclin B1 (ccnb1) mRNAs were identified at animal pole (arrow) after probe labelling with fluorescein-labelled antisense riboprobes and Deleted AZoospermia-Like (dazl) transcripts located at vegetal pole (arrowhead) after probe labelling with digoxigenin-labelled antisense riboprobes. The hybridization signal is coloured red with fluorescein and dark brown to blue with digoxigenin. B, The animal pole localisation of transcripts similar to rhamnose-binding lectin at the fully-grown follicle stage (stage IV) was detected with digoxigenin-labelled antisense riboprobes. C, No staining signal was observed using sense riboprobes of transcripts similar to rhamnose-binding lectin. D-H, Stage-specific polarised distribution of metallothionein 2 (mt2) short transcript isoform in stage IB (D, E), II (F), early stage III (G), and stage IV (H) follicles. The met2 hybridization signal detected at early stages was widely distributed in the ooplasma, whereas it concentrated at the animal pole from early stage III to the end of vitellogenesis. For stages III and IV, the animal pole, oriented toward top of page, is indicated by an arrow. Scale bar = 100 μm.
|

