- Title
-
Mutations affecting development of the notochord in zebrafish
- Authors
- Stemple, D.L., Solnica-Krezel, L., Zwartkruis, F., Neuhauss, S.C., Schier, A.F., Malicki, J., Stainier, D.Y., Abdelilah, S., Rangini, Z., Mountcastle-Shah, E., and Driever, W.
- Source
- Full text @ Development
|
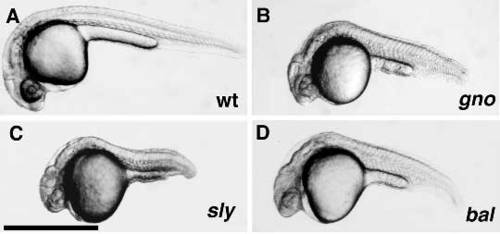
Notochord differentiation mutants at 24 hpf. Micrographs of a 24 hpf wild-type (A) embryo and 24 hpf mutant embryos of the gnom622 (B), slym86 (C), and balm190 (D) loci. Scale bar, 1 μm. PHENOTYPE:
|
|
Chordamesoderm and notochord phenotypes of boz mutants. DIC micrographs of a (A) wild-type (arrowheads indicate sides of the chordamesoderm) and (B) boz mutant embryo at the 10-somite stage demonstrate the absence of chordamesoderm in bozm168 mutant embryos. In (A) the chordamesoderm is shown bisecting the paired somites. The chordamesoderm is missing in the bozm168 mutant embryo (B) resulting in the lateral fusion of somites. (C,C′) At 24 hpf a vacuolated notochord normally seen in wild-type embryos (C; between arrowheads) is completely missing in the boz mutant embryo (C′). (D) Variable expressivity of the phenotype is observed as some boz mutant embryos form a notochord (between the arrowheads) in the tail as seen in this 72 hpf embryo tail. Scale bars, 250 μm. PHENOTYPE:
|
|
Notochord phenotype of mutants in the sly, gup, bal, dop, sny, gno and mik loci at 24 hpf. DIC micrographs of a 24 hpf (A) wildtype embryo, and 24 hpf (B) slym466, (C) gupm189, (D) balm190, (E) dopm341, (F) snym456, (G) gnom622, and (H) mikm218 mutant embryos demonstrate the failure of mutant notochords to differentiate normally and form vacuolated cells. Scale bar, 250 μm. PHENOTYPE:
|
|
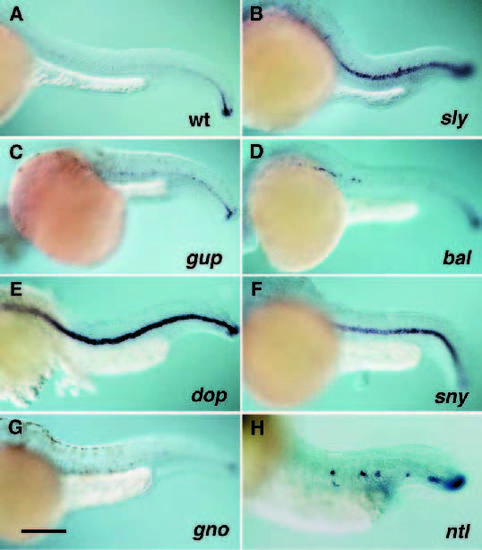
Brachyury expression in mutants in the sly, gup, bal, dop, sny, gno and ntl loci. Shown are DIC micrographs of a 32 hpf (A) wildtype embryo, and (B) slym466, (C) gupm189, (D) balm190, (E) dopm341, (F) snym456, and (G) gnom622 mutant embryos and a 24 hpf (H) ntlm550 mutant embryo stained by in situ hybridization with antisense zebrafish Brachyury RNA. Scale bar, 250 μm. |
|
col2a1, shh, and Engrailed expression in mutants in the sly, gup, bal, dop, sny, and gno loci. Shown are DIC micrographs of (A-C) wildtype embryos, and (D-F) slym466, (G-I) gupm189, (J-L) balm190, (M-O) dopm341, (P-R) snym456, and (S-U) gnom622 mutant embryos stained by in situ hybridization with anti-sense zebrafish col2a1 (A,D,G,J,M,P,S), shh (B,E,H,K,N,Q,T) RNA or with monoclonal anti-Engrailed antibody 4D9 (C,F,I,L,O,R,U). Embryos stained for col2a1 are at 32 hpf and embryos stained for shh or Engrailed are at 24 hpf. Scale bars, 250 μm. |
|
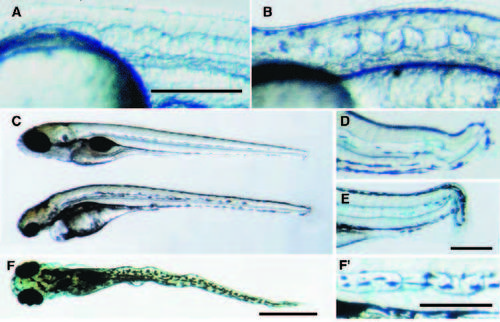
Notochord phenotype of mutants in the not, chg, mib, snw, and mgt loci (A) The notm128 phenotype first becomes apparent at 48 hpf, manifest as regions of degeneration in the notochord (middle) or the notochord and muscle (lower). (B) Wild-type 48 hour notochord under DIC optics and (C) a notm128 mutant notchord displays abnormal cellular morphology and regions of degeneration. (D) The phenotype of chgm275 mutants becomes apparent by 24 hpf. By 48 hpf the notochord cellular morphology is abnormal throughout the axis. Shown is an anterior notochord from a 48 hpf wild-type embryo (E) and a corresponding region of a chgm275 mutant notochord (F). (G) Shown is a mibm132 mutant (lower) and a wild-type sibling (upper) at 24 hpf. (H) A 24 hpf wild-type tail and (I) mibm132 mutant tail show abnormal morphology of cells in the mutant notochord. The mutation snwm454 combines two phenotypes, regions of disrupted notochord with an overall failure to form melanophores properly. (J) In a 48 hpf snwm454 mutant (lower) the lack of melanophores is especially apparent on the yolk sac and in the head. The xanthophores and iridophores and the retinal pigmented epithelium are apparently unaffected. (K) A wild-type 28 hpf notochord displays the normal scalloped notochord cell morphology. (L) Regions of the notochord in snw mutants display an unusual phenotype in which rounded cells are present instead of the normally scalloped vacuolated notochord cells. (M) By 120 hpf mgtm635 mutant embryos (lower) are severly shorter than wild-type. Additionally, the mutants have abnormal craniofacial development including reduced branchial arch development. (N) A 32 hpf wild-type notochord and (O) a mgtm635 notochord with morphologically abnormal spherical cells. Scale bars, 250 μm (A,D,G,H,J,M); 50 μm (B,E,K,N). |
|
Short axis and notochord phenotype of mutants in the pro, sno, git, and drb loci (A) Shown is a mutant (lower) at the prom672 locus at 120 hpf with a wild-type sibling (upper). Scale bar = 1mm. (B) A higher magnification image of the pro mutants shows regions of disruption in the notochord (black arrowheads). (C) A mutant (lower) in the snom563 locus at 72 hpf with a wild-type sibling (upper). (D) A mutant (lower) in the gitm342 locus at 48 hpf with a wild-type sibling (upper). (E) A mutant (lower) in the drbm759 locus at 72 hpf with a wild-type sibling (upper). Scale bars, 250 μm (B-E). |
|
Notochord shape phenotype of mutants in the gul, lev, bup, tri and kny loci A wavy notochord phenotype by 28 hpf characterizes the two mutations gulm208 (A) and levm531 (B). (C) By 72 hpf gulm208 mutants have a straight notchord and a severe head malformation (bottom). (F, F′) By contrast, by 96 hpf in levm531 mutants the wavy notochord phenotype is exacerbated and shows both lateral (F) and dorsal-ventral flexures (F′). The gastrulation mutations trim144 (D) and knym119 (E) exhibit a folded notochord phenotype only in the tail at 72 hpf. Scale bars, 250 μm (A,E, F′) and 1 mm (A,F). |