- Title
-
DNA methyltransferase Dnmt3ba-mediated epigenetic modulation of Integrin signaling is essential for hematopoietic stem and progenitor cell development
- Authors
- Ai, K., Wu, Y., Liang, G., Kong, H., Yang, X., Li, N., Liu, Z., Dong, Y., Xu, J., Zhang, L., Chen, X., Fu, Y., Wang, L., Li, L.
- Source
- Full text @ Commun Biol
|
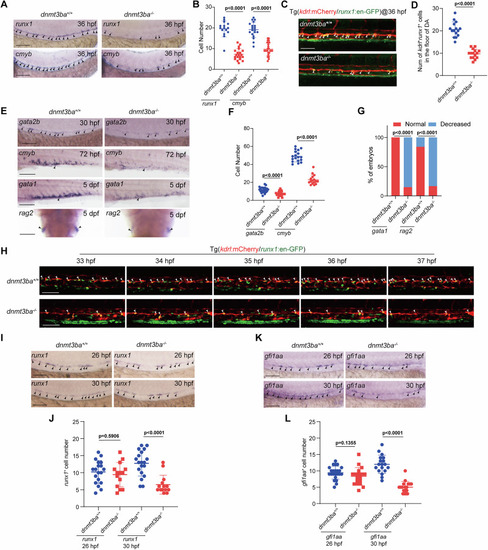
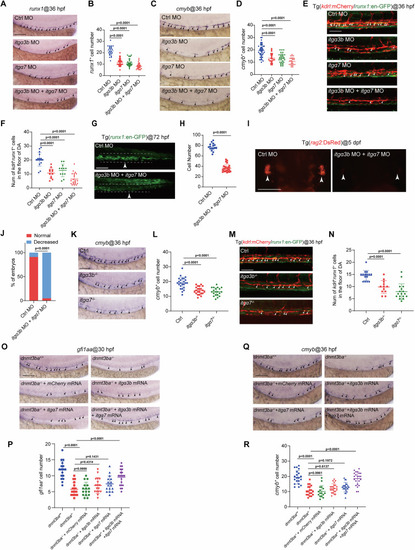
Dnmt3ba is required for EHT and HSPC development. |
|
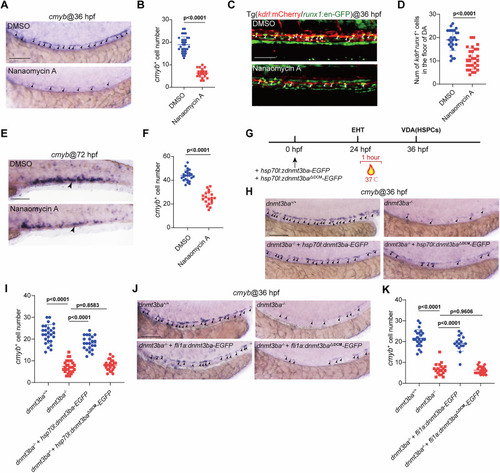
The DNA methyltransferase activity of Dnmt3ba is required for HSPC development. |
|
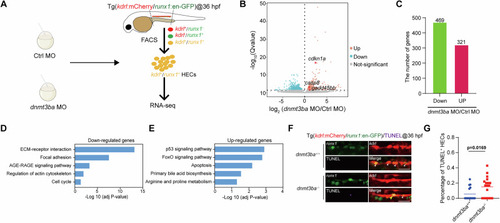
Transcriptomic analysis of |
|
|
|
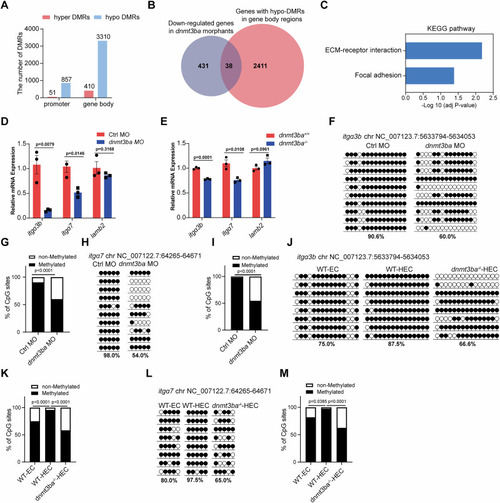
HSPC development is impaired in |
|
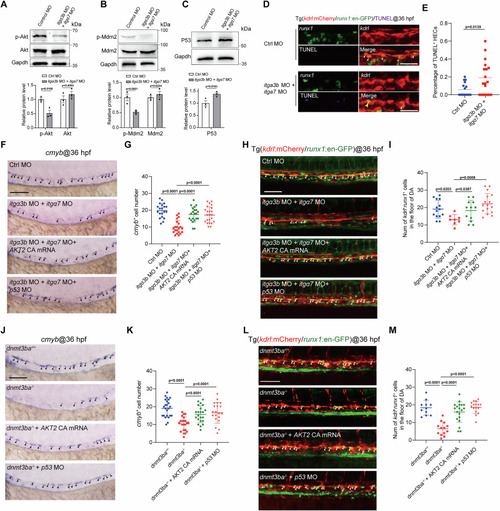
Integrin signaling controls HSPC development through regulating the Akt and P53 pathways. |
|
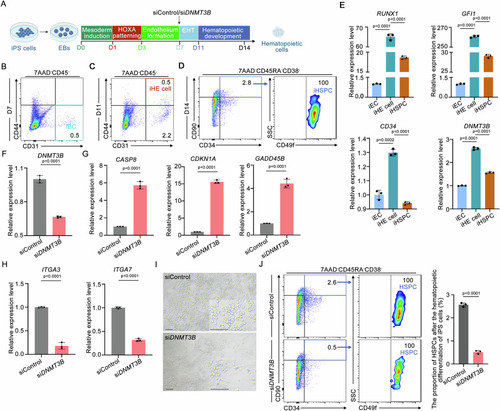
DNMT3B knockdown impairs the iHSPCs generation during hematopoietic differentiation from human iPS cells in vitro. |