- Title
-
Identification of Immune Hub Genes in Obese Postmenopausal Women Using Microarray and Single-Cell RNA Seq Data
- Authors
- Zhang, F.R., Lu, X., Li, J.L., Li, Y.X., Pang, W.W., Wang, N., Liu, K., Zhang, Q.Q., Deng, Y., Zeng, Q., Qu, X.C., Chen, X.D., Deng, H.W., Tan, L.J.
- Source
- Full text @ Genes (Basel)
|
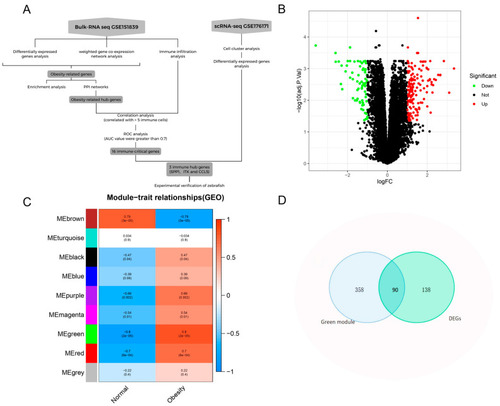
Identification of obesity-related genes. ( |
|
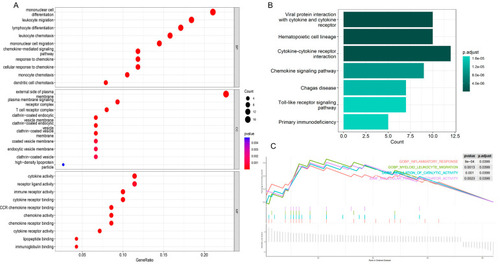
Enrichment analysis of obesity-related genes. ( |
|
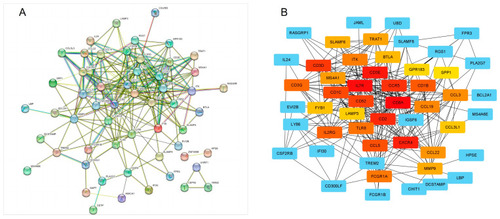
PPI network of obesity-related genes. ( |
|
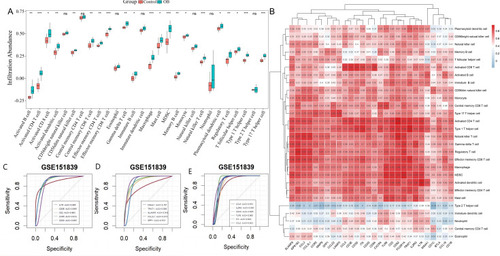
Immune infiltration analysis. ( |
|
Single-cell RNA sequencing analysis of adipose tissue immune populations. ( |
|
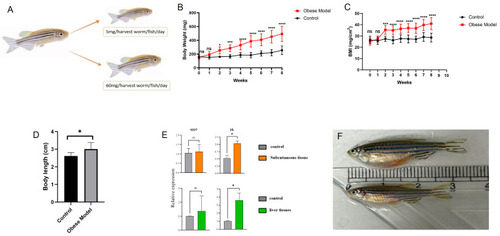
Verification of immune hub genes in zebrafish. ( |