- Title
-
Benchmarking metabolic RNA labeling techniques for high-throughput single-cell RNA sequencing
- Authors
- Zhang, X., Peng, M., Zhu, J., Zhai, X., Wei, C., Jiao, H., Wu, Z., Huang, S., Liu, M., Li, W., Yang, W., Miao, K., Xu, Q., Chen, L., Hu, P.
- Source
- Full text @ Nat. Commun.
|
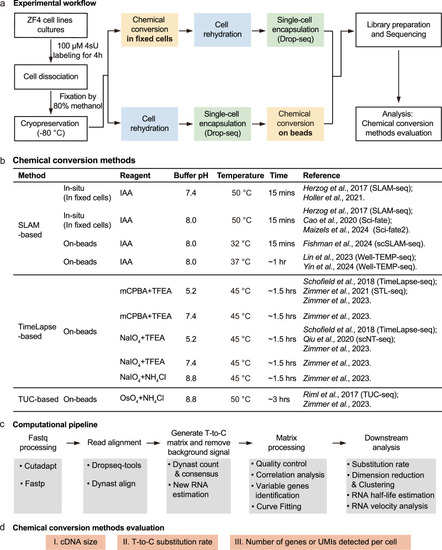
Experimental design for benchmarking chemical conversion methods. |
|
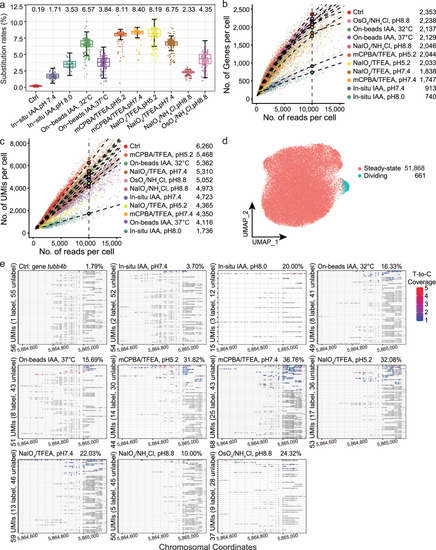
Comparison and evaluation of ten chemical conversion methods using ZF4 cells. |
|
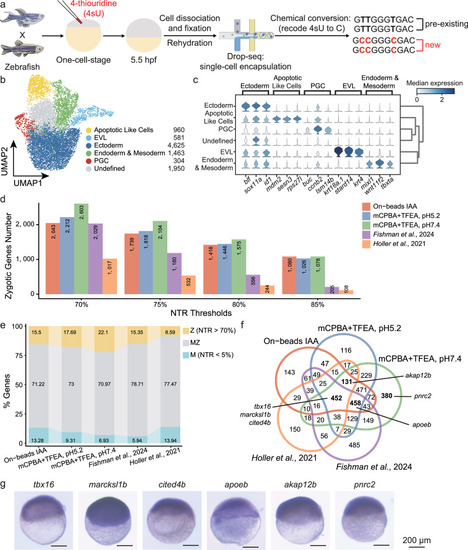
Identification of zygotically activated transcripts in zebrafish embryogenesis using improved chemical conversion methods. |
|
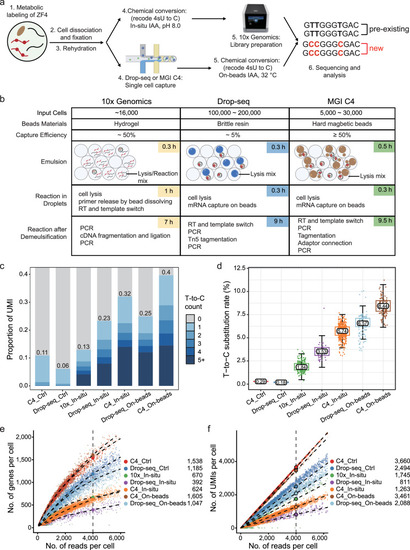
Comparison between 10× Genomics, Drop-seq and MGI C4 high-throughput single-cell platforms. |