|
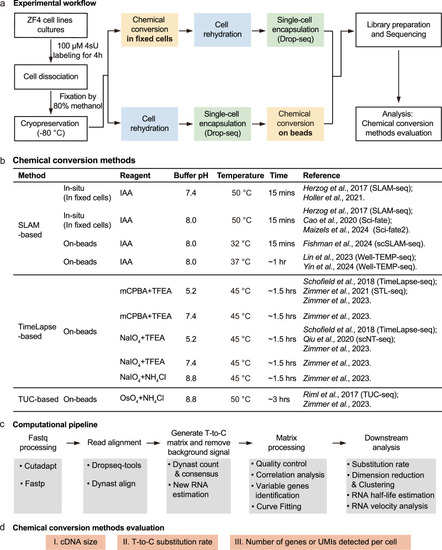
Experimental design for benchmarking chemical conversion methods. a Workflow for high-throughput scRNA-seq using metabolic labeling in ZF4 cells. ZF4 cells were labeled with 4-thiouridine (4sU, 100 μM), followed by cell dissociation and fixation. Chemical conversion was performed either before or after single-cell encapsulation on the Drop-seq platform. Newly synthesized transcripts were detected via sequencing by identifying chemical-induced T-to-C substitutions. b Summary of the ten chemical conversion methods evaluated in this study, including key parameters such as the main reagent, buffer pH, temperature, reaction time, and relevant references. “In-situ” refers to chemical conversion occurring within intactly fixed cells, while “on-beads” indicates that the chemical conversion occurs after mRNA is released from the cells and captured on beads. IAA iodoacetamide, mCPBA meta-chloroperoxy-benzoic acid, TFEA 2,2,2-trifluoroethylamine, NaIO4 sodium periodate, NH4Cl ammonium chloride, OsO4 osmium tetroxide. c Computational pipeline for data processing, starting with fastq file pre-processing using Cutadapt and fastp, followed by read alignment with Dynast and Dropseq-tools. T-to-C substitutions were identified using Dynast, with R and Python scripts used for cell quality control, dimension reduction, new transcript identification, and RNA velocity analysis (see details in “Methods”). d Benchmarking criteria used to evaluate chemical conversion performance, focusing on cDNA size, T-to-C substitution rate, and the number of genes and unique molecular identifiers (UMIs) detected per cell.
|