- Title
-
Impacts of ribosomal RNA sequence variation on gene expression and phenotype
- Authors
- Welfer, G.A., Brady, R.A., Natchiar, S.K., Watson, Z.L., Rundlet, E.J., Alejo, J.L., Singh, A.P., Mishra, N.K., Altman, R.B., Blanchard, S.C.
- Source
- Full text @ Phil. Trans. Roy. Soc. Lond., Series B
|
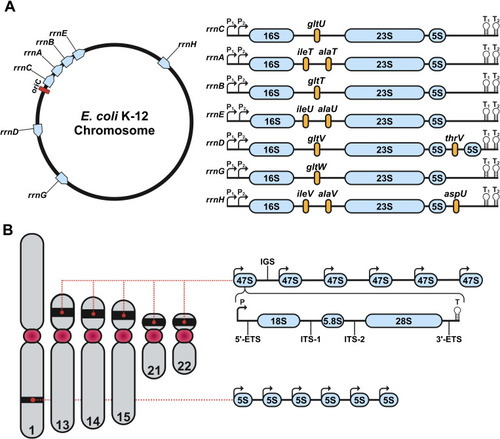
Genomic organization of rDNA in |
|
Model of ribosome heterogeneity in |
|
Variant ribosomes exhibit altered drug sensitivity. (A,B) BIOLOG phenotypic screening comparing the growth of Δ7prrn-BBB (red) and Δ7prrn-HBB (cyan) in the presence of (A) tetracycline and (B) oxytetracycline. Panels A and B are modified from Kurylo |
|
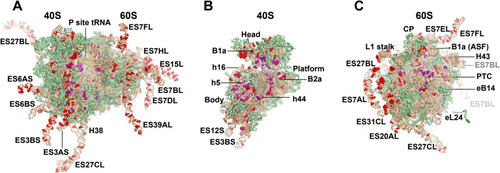
High-frequency rRNA variants detected in humans. (A) 80S human ribosome with modeled expansion segments labelled at high-frequency allelic variant positions (>20%). (B) 40S small subunit of human ribosome with high-frequency variant alleles. (C) 60S large subunit of human ribosome with variant high-frequency variant alleles. Key structural features of the ribosome are indicated. Key: rRNA, tan; RP, green; variant nucleotide, red; posttranscriptional modifications, purple. Model based on PDB: 8G5Y. Gray labels of expansion segments indicate their relative distance from the viewers perspective. |
|
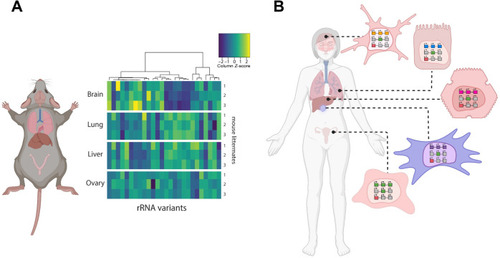
Tissue-specific expression of rRNA variants. (A) Experimentally observed variance in rRNA across different mouse tissues. This panel is reprinted from a previous publication [ |