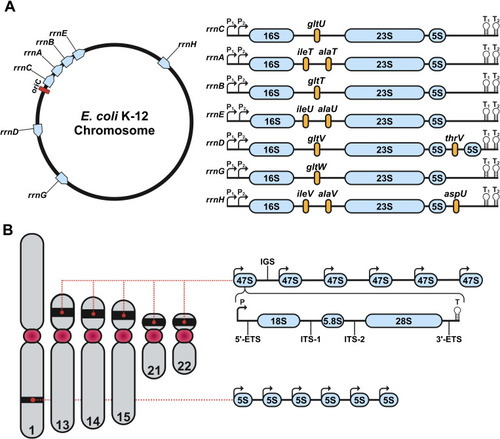
Genomic organization of rDNA in E. coli and human. (A) Chromosomal map of the seven rDNA operons (rrnC, rrnA, rrnB, rrnE, rrnD, rrnG and rrnH) in E. coli. On the left, each operon’s position is shown in blue relative to the origin of replication (oriC, red). On the right appear genetic structures of the seven rDNA operons. Each rDNA operon consists of the 16S, 23S and 5S rRNA genes. rrnD contains a second 5S gene, whereas all other operons only encode a single copy of each component. The rRNA genes are separated by internally transcribed regions, as well as unique tRNAs (yellow). Transcription initiation sites are marked by P₁ and P₂, termination sites for transcription are marked by T₁ and T₂. (B) Genomic organization of the rDNA operon in the human genome. Chromosome 1 encodes tandem repeats of 5S rRNA. Chromosomes 13, 14, 15, 21 and 22 encode tandem arrays of 47S rDNA operons within their p-arms. Each human rDNA operon contains a promoter (P), a 5’ externally transcribed region (5’-ETS), the 18S rRNA, internally transcribed region 1 (ITS-1), the 5.8S rRNA, internally transcribed region 2 (ITS-2), the 28S rRNA, the 3’ externally transcribed region (3’-ETS) and a transcriptional termination motif (T). Furthermore, each operon is separated by an intergenic spacing region (IGS). Created with BioRender.com.
|

