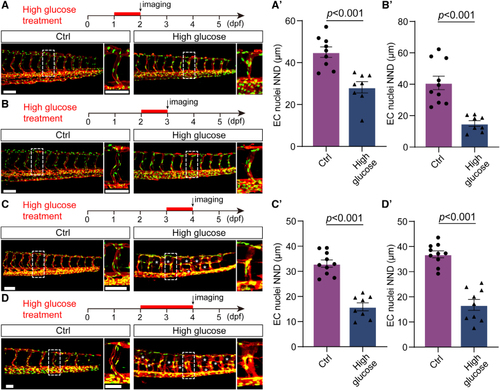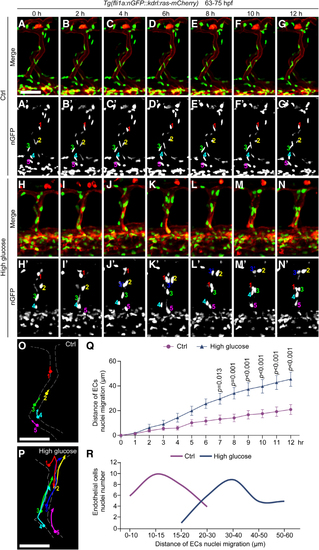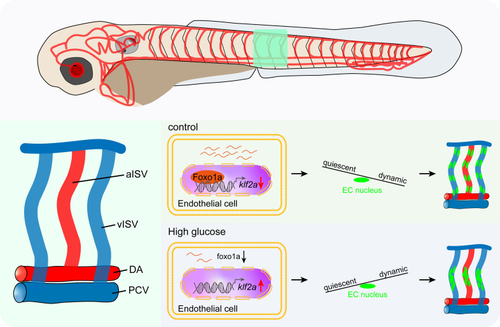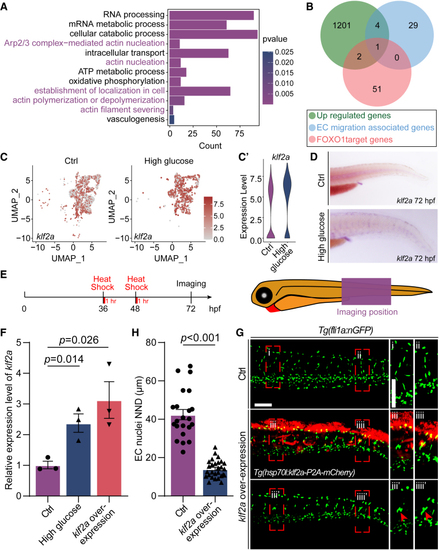
Klf2a (Krüppel-like transcription factor 2a) was involved in the nuclei aggregation induced by high glucose treatment. A, GO analysis of 1201 upregulated genes in arterial and capillary endothelial cells (ECs). B, Venn diagram showing the overlap between FOXO1 (forkhead box protein O1) target genes, EC migration–associated genes, and significantly upregulated genes. C, The feature plot of klf2a of control (Ctrl) and high glucose group in arterial and capillary ECs. C′, The violin plot of klf2a of Ctrl and high glucose group in arterial and capillary ECs. D, Whole-mount in situ hybridization analysis of klf2a in Ctrl and high glucose–treated embryos. E, Schematic diagram of heat shock treatment and confocal imaging region. F, Real-time PCR analysis of klf2a expression in Ctrl, high glucose–treated, and klf2a overexpressed embryos. Each dot represents data from an independent experiment (n=3). t test. G, Confocal imaging analysis of Ctrl embryos and Tg(hsp70l:klf2a-P2A-mCherry) embryos at 72 hours post-fertilization (hpf). The right panels (i, ii, iii, iiii, iii', iiii') are the magnifications of the red dotted boxes. Arrowheads indicate the EC nuclei. H, Statistics of the EC nuclei nearest neighbor distance (NND) in Ctrl (n=22) and Tg(hsp70l:klf2a-P2A-mCherry) embryos (n=28). t test. Scale bars, 100 µm. Arp2/3 indicates actin-related proteins-2/3; GO, gene ontology; PCR, polymerase chain reaction; and UMAP, uniform manifold approximation and projection.
|