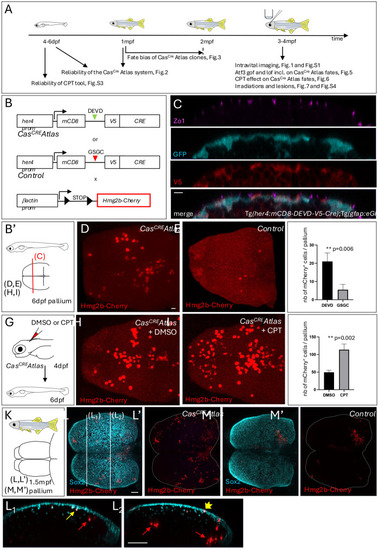
CasCREAtlas reveals non-apoptotic caspase events during pallium development and homeostasis. (A) Experiments carried out in the present study shown in relation to the zebrafish lifetime. (B) Schematics of the transgenic lines used. CasCREAtlas combines an NP/NSC-specific driver line (top and middle) expressing Cre upon Cas3*/Cas7* cleavage and a reporter line (bottom) expressing hmg2bmCherry upon Cre recombination. mCD8, membrane anchor; V5, antigen segment (tag); DEVD, canonical Cas3*/Cas7* cleavage site; GSGC, non-productive cleavage site; her4 prom, her4 regulatory elements. The Tg(her4:mCD8-DEVD-V5-Cre);Tg(βact;lox-stop-lox-hmg2bmCherry) background is referred to as CasCREAtlas, and Tg(her4:mCD8-GSGC-V5-Cre);Tg(βact;lox-stop-lox-hmg2bmCherry) as ‘control’. All analyses were conducted in double heterozygotes. (B′) Schematic of analyses in cross-sectioned or whole-mount pallia at 6 dpf. (C) Expression of the CasCRE driver is confined to gfap:eGFP-positive cells. Pallial cross-sections of Tg(her4:mCD8-DEVD-V5-Cre);Tg(gfap;eGFP) larvae at 6 dpf with IHC for GFP (NP/NSC), Zo1 (tight junctions, delimiting apical surfaces) and V5 (transgene tag), showing V5 (membrane anchored) and GFP in the same cells. (D-F) Hmg2bmCherry whole-mount IHC on CasCREAtlas (D) and control (E) pallia at 6 dpf (dorsal views, z-projections, anterior left); quantification of positive cells (F) (CasCREAtlas: n=4 brains; control: n=10 brains; Mann–Whitney test). (G-J) CasCREAtlas activity is induced by camptothecin (CPT). (G) Schematic of the experiment: CPT (20 µM) or DMSO was injected into the neural tube ventricle in CasCREAtlas larvae at 4 dpf; Hmg2bmCherry was analyzed at 6 dpf. (H-J) Hmg2bmCherry whole-mount IHC at 6 dpf (dorsal views, z-projections, anterior left) and quantification (J) (DMSO and CPT: n=6 brains each; Mann–Whitney test). (K-M′) Hmg2bmCherry expression in CasCREAtlas (L,L′) and control (M,M′) pallia at 1.5 mpf. (K) Schematic of whole-mount pallia at 1.5 mpf. (L-M′) Whole-mount IHC for Sox2 (NPs, NSCs and some freshly born neurons) and Hmg2bmCherry (dorsal views, z-projections, anterior left). L1 and L2 show cross-sections as indicated in L, dorsal up. Red arrows indicate Sox2neg neurons, thin yellow arrow Sox2pos neurons, and short yellow arrow a Sox2pos NSC. Scale bars: 10 µm (C-E,H,I); 50 µm (L-M′,L1,L2).
|