- Title
-
Cohesin rad21 mutation dysregulates erythropoiesis and granulopoiesis output within the whole kidney marrow of adult zebrafish
- Authors
- Gimenez, G., Kalev-Zylinska, M.L., Morison, I., Bohlander, S.K., Horsfield, J.A., Antony, J.
- Source
- Full text @ Am. J. Physiol. Cell Physiol.
|
WKM of rad21+/− zebrafish show transcriptional dysregulation and altered erythroid and granulocytic outputs. A: schematic of experimental process. B: principal component analyses of the WT and rad21+/− WKM bulk RNA sequencing. C: WKM bulk RNA sequencing- MA plot showing DEGs identified in the comparison of rad21+/− versus WT. Red and blue denote significant (adjusted P ≤ 0.05) DEGs in rad21+/− compared with WT. D: representative MayGrunwald/Giemsa staining of WKM cytospins. Blue arrows denote examples of maturing granulocytes and orange arrows denote examples of orthochromatic erythroid cells. E: the percentage of different hematopoietic cells identified in the WKM cytospins. F: ratio of myeloid (includes maturing granulocytes, myeloid precursors, and monocytes/macrophages) or maturing granulocytic cells relative to erythroid cells in the WKM. Data in E and F were obtained by counting 1,000–2,000 cells in WKM cytospins prepared from each zebrafish. Error bars represent means ± SE (n = 6 per genotype). Significance in E and F was determined using Student’s t test. *P < 0.05. E, erythroid cells (includes basophilic, polychromatic, and orthochromatic erythroid); Gr, maturing granulocytic cells (include promyelocytes, myelocytes, metamyelocytes, bands, segmented forms, and eosinophils/basophils); L, lymphoid; M, monocytes/macrophages; MP, myeloid precursors (granulocytic or monocytic); T, thrombocytes; UP, undifferentiated precursors; WKM, whole kidney marrow; WT, wild type. PHENOTYPE:
|
|
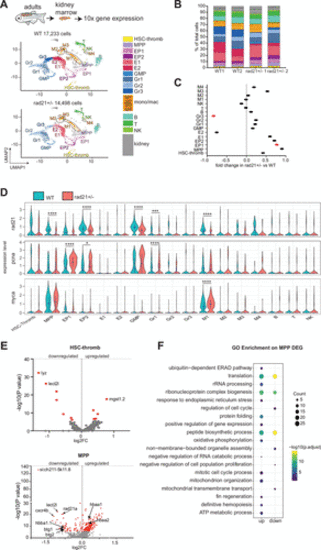
Single-cell RNA sequencing reveals the hematopoietic composition in WKM of adult wild type and rad21+/− zebrafish. A: uniform manifold projection (UMAP) showing the 22 cell clusters identified in scRNA-seq of WKM cells from wild type (WT) and rad21+/− adult zebrafish. B: stacked bar plot showing the percentage of annotated cell population within each genotype per replicate. WT1: 8,125 cells, WT2: 9,108 cells, rad21+/− 1: 6,159 cells, rad21+/− 2: 8,339 cells. C: fold change in population frequency in rad21+/− compared with WT. Red dot signifies statistically significant (P ≤ 0.05) change. Significance was determined using Student’s t test. D: violin plots of rad21, pcna, and myca expression in rad21+/− and WT. In rad21+/− versus WT comparison significance is from the scRNA-seq DESeq2 Wald test (Supplemental Table S3). *P < 0.05, ***P < 0.001, ****P < 0.0001. E: volcano plots showing DEGs identified in the comparison of rad21+/− versus WT in HSC-thromb and MPP clusters. Red dot signifies statistically significant (adjusted P ≤ 0.05) DEGs. F: GO enrichment analyses on up and down significant DEGs identified in MPP. Pathway analyses were performed using ClusterProfiler R package. Complete list of pathways and associated genes is provided in Supplemental Table S4. B, B-lymphocyte cells; E, erythroid; EP, erythroid progenitors; GMP, granulocytes monocyte progenitors; Gr, granulocytes; HSC, hematopoietic stem cell; M, monocytes/macrophages; MPP, multipotent progenitors; NK, natural killer; T, T-lymphocyte cells; thromb, thrombocytes; WKM, whole kidney marrow. |
|
Erythroid differentiation program is altered in rad21+/− WKM. A: UMAP of scRNA-seq data showing erythroid cell clusters and Monocle 3 pseudotime trajectory within the erythroid lineage starting from MPP. B: violin plots showing the distribution of the pseudotime values for MPP and erythroid cells. Indicated in blue dots are the median pseudotime values for each cluster. Significance was determined using the Wilcoxon rank-sum test. ****P < 0.0001. C: volcano plot showing DEGs identified from comparison of rad21+/− versus WT in E1 and E2. Red dot signifies statistically significant (P < 0.05) DEGs. D: violin plots showing the expression of erythroid associated genes. Significance is from the scRNA-seq DESeq2 Wald test. **P < 0.01, ***P < 0.001, ****P < 0.0001 (Supplemental Table S3). E: GO enrichment (using Metascape) on the significant DEGs (P ≤ 0.05) identified in M1 from comparison of rad21+/− versus WT cells. Complete list of pathways and the associated DEGs are provided in Supplemental Table S5. F: volcano plot showing DEGs identified in M1 and M3 in comparison of rad21+/− versus WT. Red dot signifies statistically significant (adjusted P ≤ 0.05) DEGs. Shown in green are iron storage genes and in purple are the erythrocyte and cytokine genes. WKM, whole kidney marrow; WT, wild type. |
|
Granulopoiesis is impaired in rad21+/− WKM but granulocytic maturation is not overtly disrupted. A: UMAP of scRNA-seq data showing the granulocyte cell clusters and Monocle 3 pseudotime trajectory within the granulocytic lineage starting from MPP. B: violin plots showing distribution of pseudotime values for MPP and granulocytic cells. Indicated in blue dots are the median pseudotime values for each cluster. Significance was determined using the Wilcoxon rank-sum test. *P < 0.05, **P < 0.01. C: volcano plot showing DEGs identified in GMP and Gr3 in comparison of rad21+/− versus WT. Red dot signifies statistically significant (adjusted P ≤ 0.05) DEGs. Shown in green are some of the genes associated with stress. D: violin plots showing the expression of granulocyte marker genes. Significance is from scRNA-seq DESeq2 Wald test. **P < 0.01, ***P < 0.001, ****P < 0.0001. WKM, whole kidney marrow; WT, wild type. |