- Title
-
Generation of Zebrafish Maternal Mutants via Oocyte-Specific Knockout System
- Authors
- Zhang, C., Wei, W., Lu, T., Zhang, Y., Li, J., Wang, J., Chen, A., Wen, F., Shao, M.
- Source
- Full text @ Bio Protoc
|
Main steps of this protocol |
|
Example output from CRISPRScan. Zebrafish-Danio rerio, Cas9-NGG, Gene, in vitro T7 promoter, and 4 mismatches should be chosen, and the gene symbol is then submitted to “Get sgRNAs”. This protocol uses Cas9 for genome editing, so we select Cas9-NGG to specify the prediction of sgRNA for Cas9. Allowing for “4 mismatches” enables the sgRNA site to tolerate up to four mismatches in off-target sequences. This criterion helps to identify a broader range of potential target sites for consideration. |
|
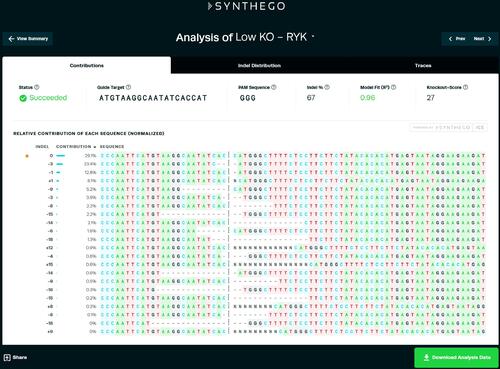
Example output of ICE analysis. We consider the value of “indel %” as the efficiency of the sgRNA. |
|
Schematics illustrating the construction of U6 sgRNA expression vector. All U6 vectors have a U6 promoter and a sgRNA scaffold. Digestion of BsmBI makes U6 vectors expose the overhangs recognizing the annealed sgRNA primer. Digestion of PstI and SalI can disrupt the vector undigested by BsmBI to improve positive cloning. |
|
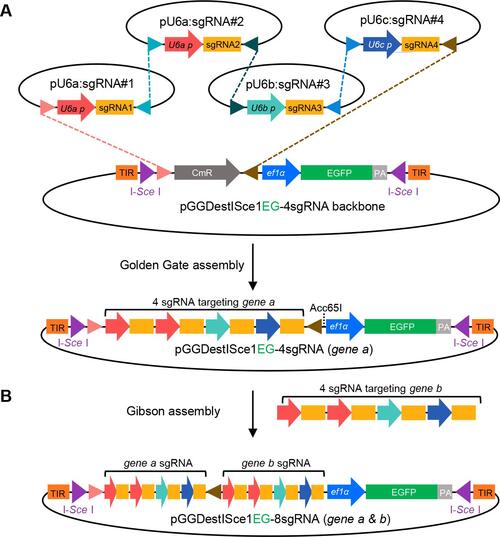
Golden Gate cloning of tandem sgRNA expression vector and Gibson assembly of double-genes sgRNA expression cassettes. A. BsaI digestion leaves specific overhangs, which are designed for tandem ligation into pGGDestISceIEG-4sgRNA. B. The amplified sgRNA expression cassette targeting gene b can be cloned into the vector of gene a through Gibson assembly. |
|
Representative electrophoresis result of I-Sce efficiency test. I-Sce I digestion can split the original plasmid into two fragments. No original plasmids should be found when the I-Sce1 works well. |
|
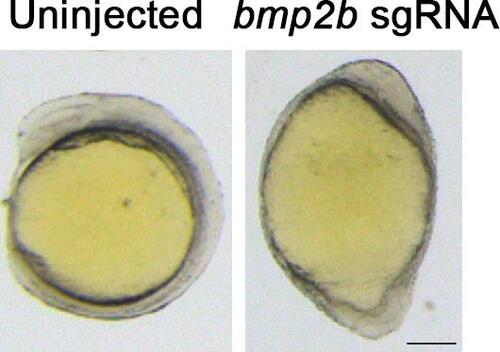
Representative images of normal and dorsalized embryos at 12 hpf. Scale bar: 200 μm |
|
Illustration of procedures of phenotyping and genotyping individual embryos [11,12,18] |
|
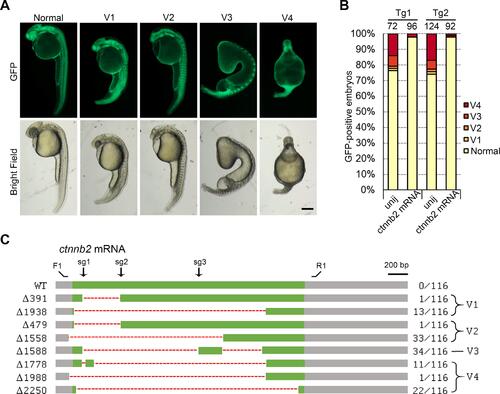
Generation of ctnnb2 maternal mutants using this protocol. These data were reproduced from our previous publication [12]. A. Phenotypes of the GFP-positive embryos expressing three sgRNAs targeting ctnnb2. B. Injection of wild-type ctnnb2-myc mRNA at the 1-cell stage could efficiently rescue the ventralized phenotypes. C. Schematics shows the mutation types by examining colonies. |
|
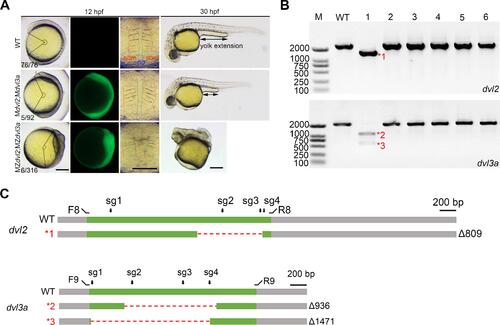
Generation of dvl2 and dvl3a maternal mutants using this protocol. These data were reproduced from our previous publication [11]. A. Phenotypes of wild-type, Mdvl2;Mdvl3a and MZdvl2;MZdvl3a embryos at 12 hpf and 30 hpf. B. Gel analysis after amplifying the coding sequence of dvl2 and dvl3a. C. Large deletions happened both in dvl2 and dvl3a coding sequences. |