- Title
-
Naa80 is required for actin N-terminal acetylation and normal hearing in zebrafish
- Authors
- Ree, R., Lin, S.J., Sti Dahl, L.O., Huang, K., Petree, C., Varshney, G.K., Arnesen, T.
- Source
- Full text @ Life Sci Alliance
|
Actin N-terminal maturation scheme and the in vitro acetyltransferase activity of |
|
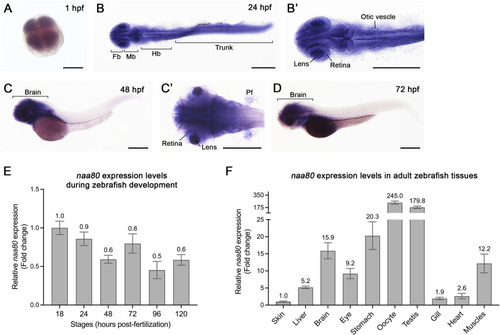
The spatiotemporal expression of |
|
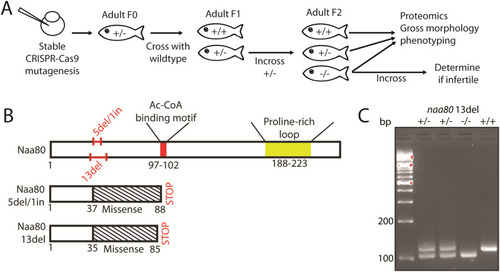
Generation of stable mutant |
|
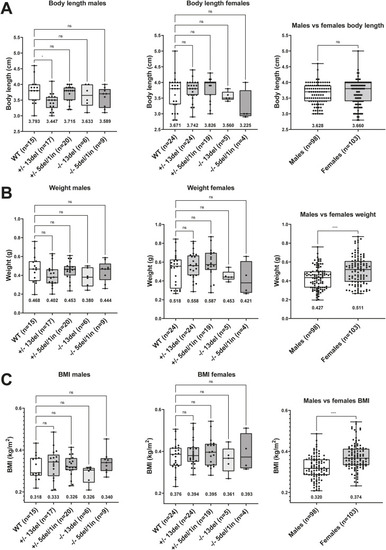
Body length and weight are not affected by Adult zebrafish with the indicated genotype were weighed and measured from rostrum to caudal tip. A one-way ANOVA test followed up with a Dunnett’s test was performed for the genotype comparison segregated by sex, where the +/+ values was selected as the control mean. *: PHENOTYPE:
|
|
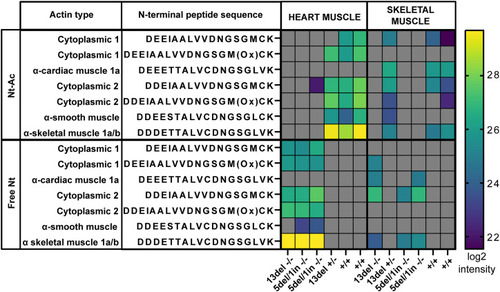
Defective in vivo actin Nt-acetylation in Mass spectrometry measurements of actin N-terminal peptides in cardiac muscle or skeletal muscle from the indicated genotypes. Log2 intensity of the peptides is reported. Gray: not identified in the sample. |
|
Zebrafish |
|
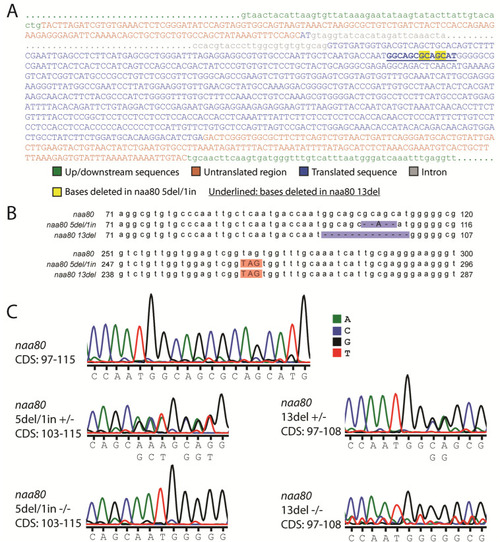
Sequences of |
|
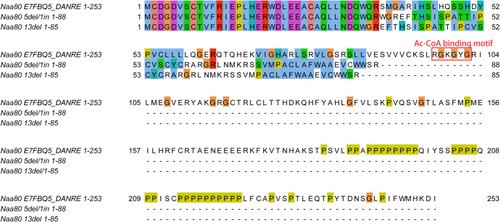
Alignment of predicted protein sequences of Naa80, and the gene products of the |