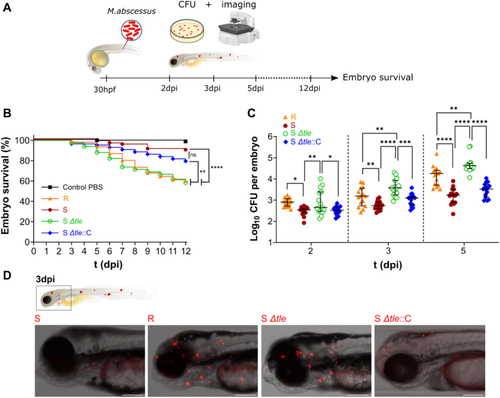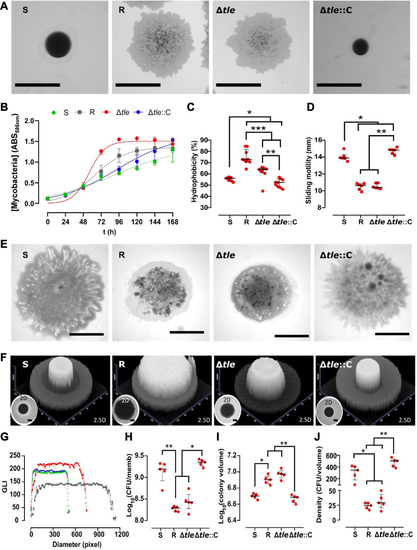
Deletion of tle in M. abscessus alters colony morphology, hydrophobicity, sliding motility, and biofilm-colony development.A, knock-out of tle is associated with a switch from an S to an R-like morphotype. The scale bar represents 1 mm. B, the deletion of tle is associated with a higher growth rate. The experiment was performed using two biological replicates with six technical replicates per biological replicate. Deletion of tle modifies hydrophobicity (C) and the sliding motility (D and E). The hydrophobicity experiment was performed using three biological replicates with three technical replicates per biological replicate. The sliding motility experiment was performed using three biological replicates with two technical replicates per biological replicate. The black scale bar indicates 5 mm. F, representative 2- and 2.5-dimensional pictures of colony-biofilms on biofilm-supporting membranes after 5 days of incubation. The absence of tle alters the biofilm-colony growth (F) (the scale bar represents 2 mm) and the biofilm-colony profile (G) (green, dark grey, red, and blue dots represent the profiles of S, R, Δtle and Δtle::C, respectively). The CFU per membrane are shown in (H), the colony volume in (I), and the colony density in (J). This experiment was performed using five biological replicates. The error bar denotes the interquartile range. ∗: p < 0.05, ∗∗: p < 0.01, and ∗∗∗: p < 0.001. CFU, colony-forming units.
|