- Title
-
A zebrafish gene with sequence similarities to human uromodulin and GP2 displays extensive evolutionary diversification among teleost and confers resistance to bacterial infection
- Authors
- Naruoka, S., Sakata, S., Kawabata, S., Hashiguchi, Y., Daikoku, E., Sakaguchi, S., Okazaki, F., Yoshikawa, K., Rawls, J.F., Nakano, T., Hirose, Y., Ono, F.
- Source
- Full text @ Heliyon
|
A. Heat map comparison of gene expression between control siblings (control) and partially paralyzed |
|
A. Amino acid sequence of |
|
A. A time course of |
|
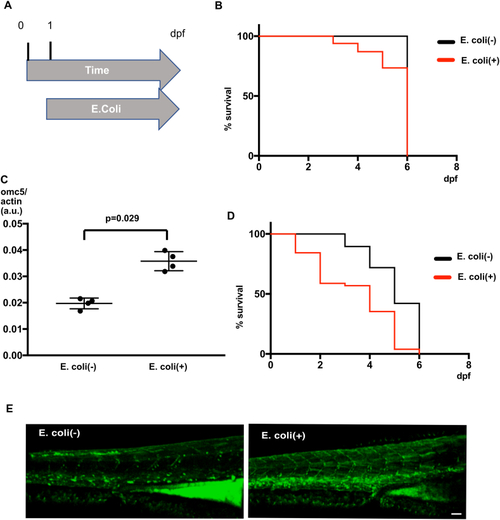
A. Diagram of |
|
A. A maximum-likelihood phylogenetic tree of the zebrafish |