- Title
-
Synthesis and Structure of Novel Phenothiazine Derivatives, and Compound Prioritization via In Silico Target Search and Screening for Cytotoxic and Cholinesterase Modulatory Activities in Liver Cancer Cells and In Vivo in Zebrafish
- Authors
- Kisla, M.M., Yaman, M., Zengin-Karadayi, F., Korkmaz, B., Bayazeid, O., Kumar, A., Peravali, R., Gunes, D., Tiryaki, R.S., Gelinci, E., Cakan-Akdogan, G., Ates-Alagoz, Z., Konu, O.
- Source
- Full text @ ACS Omega
|
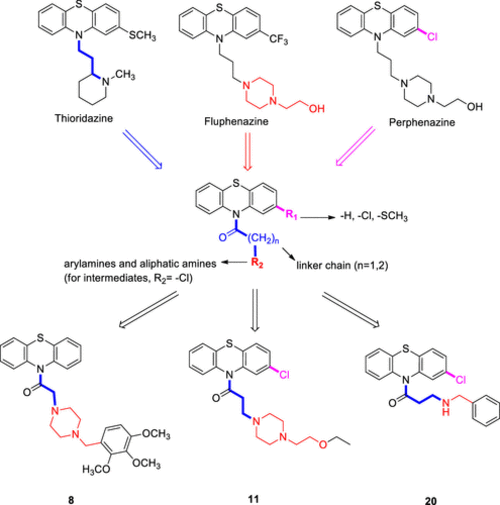
Design strategy for PTZ derivatives based on the structures of cytotoxic phenothiazines. |
|
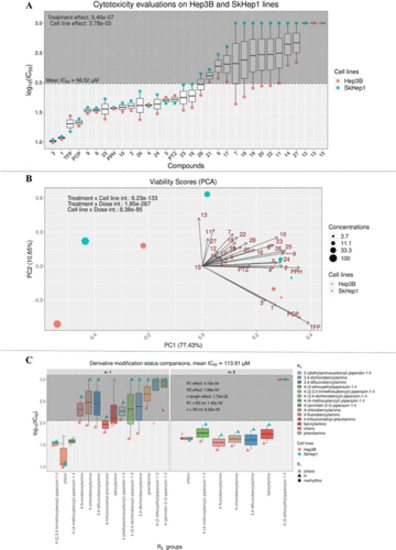
Changes in cell viability upon exposure to the derivatives in Hep3B and SkHep1 cells. (A) Known and novel derivatives’ IC50 values; (B) PCA on cell viabilities across different doses of drugs; and (C) side-chain modifications by the intermediary and novel derivatives and their influences on the IC50 levels. Significance levels (p-values) are derived from n-way ANOVA for each respective comparison in R environment. |
|
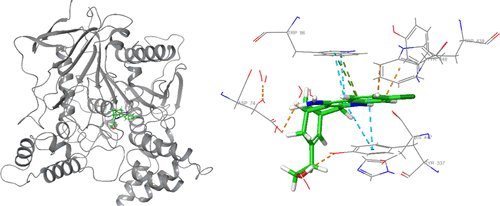
Binding mode of HUW with AChE. Several interactions with polar and hydrophobic residues were apparent. |
|
Interaction profile of HUW in the binding site (glide score = −14.76). Purple color represents H-bond interactions, whereas red lines define Pi–cation interactions. Green lines represent Pi–Pi interactions. |
|
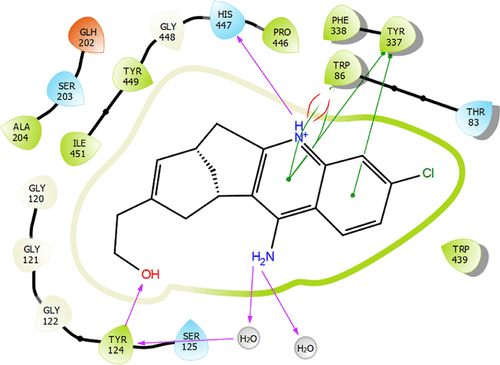
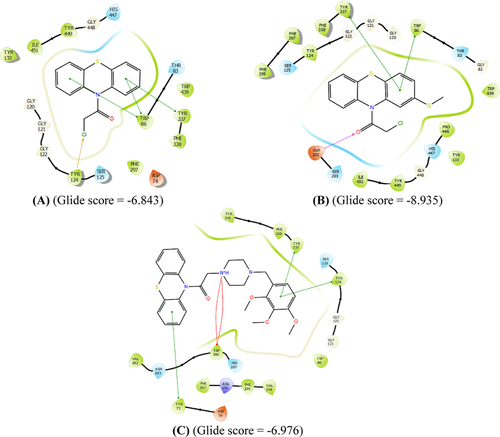
Interaction profiles of the most potent ligands 1 (A), 3 (B), and 8 (C) against HCC cell lines. Purple color represents H-bond interactions, whereas red elliptic line defines Pi-cation interaction. In addition, green lines represent Pi–Pi interactions, and gold arrow is for the halogen bond interaction. |
|
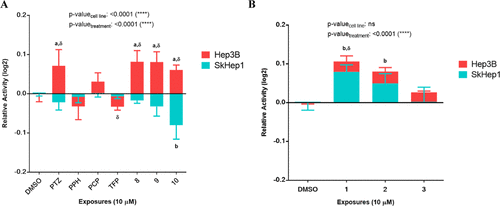
Cholinesterase activity level changes upon PTZ derivative exposures. (A) Hep3B and SkHep1 cholinesterase activity levels. (B) Cholinesterase activity levels after 24 h PTZ derivative exposures to SkHep1 and Hep3B cells. Two-way ANOVA/Dunnett’s comparisons test with respect to DMSO control (p-values: a,b ≤ 0.05) and multiple t tests/Holm-Sidak between the cell lines (p-value: δ ≤ 0.05) were applied as the statistical methods |
|
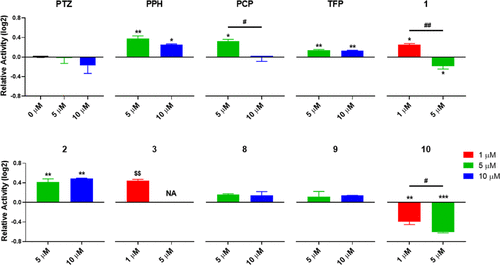
Zebrafish embryo cholinesterase activity levels after 48–120 hpf exposures: one-way ANOVA/Tukey tests with respect to DMSO control or across the applied concentrations, respectively (p-values: *,# ≤ 0.05, **,## ≤ 0.01, and ***,### ≤ 0.001), or unpaired t tests against DMSO control where total mortality was observed for the secondary groups (NAs) ($$ ≤ 0.01). |
|
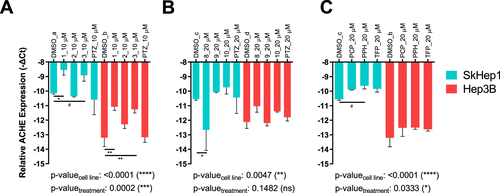
Expression of ACHE in SkHep1 and Hep3B cells, respectively, after treatment with (A) 1, 2, 3, PTZ at 10 μM; (B) 8, 9, 10, PTZ at 20 μM; and (C) PCP, PPH, TFP at 20 μM for 24 h. While the y-axis shows relative ACHE expression to TPT1 reference gene as─DeltaCt, two-way ANOVA followed by Sidak’s test was used to compare each treatment group to a batch and cell-line specific DMSO control group, indicated as DMSO_a–d. Main group tests are reported on graphs as cell line and treatment-specific p-values (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, ****: p ≤ 0.0001, and #: p ≤ 0.1). |
|
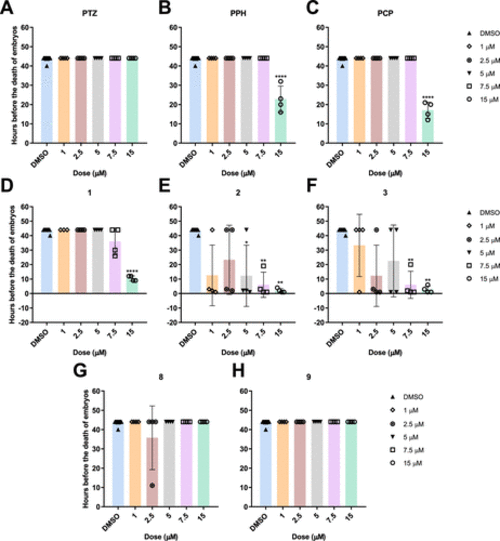
Number of hours before the death of the embryos at 9–24 hpf stage after being treated with different concentrations of known phenothiazines (A) PTZ, (B) PPH, (C) PCP, intermediate phenothiazines (D) 1, (E) 2, (F) 3, and novel phenothiazines (G) 8 and (H) 9. The statistical analysis was performed using the Kruskal–Wallis test (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, and ****: p ≤ 0.0001). |
|
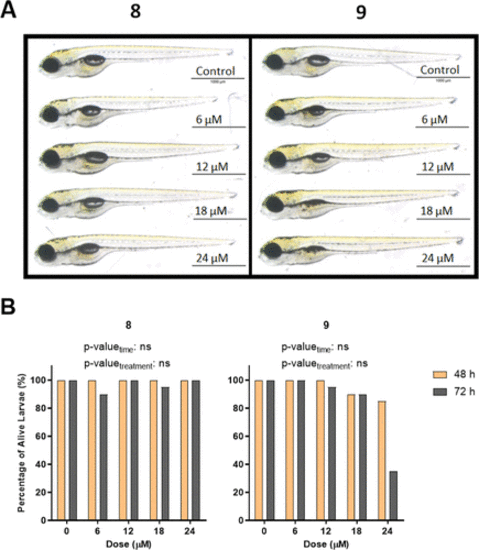
(A) Representative images of 5 dpf larvae after 72 h of exposure with novel phenothiazines 8 and 9. (B) Percentage of alive larvae after treatment with different concentrations of compounds 8 and 9 for 48 and 72 h starting from 2 dpf. The statistical analysis was performed using two-way ANOVA (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, and ****: p ≤ 0.0001). |
|
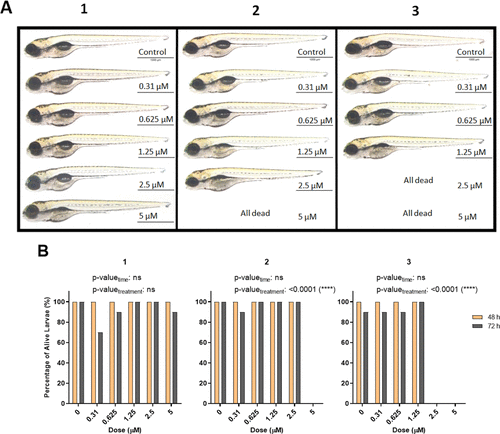
(A) Representative images of 5 dpf larvae after 72 h of exposure with intermediate phenothiazines 1, 2, and 3. (B) Percentage of alive larvae after treatment with different concentrations of compounds 1, 2, and 3 for 48 and 72 h starting from 2 dpf. The statistical analysis was performed using two-way ANOVA (*: p ≤ 0.05, **: p ≤ 0.01, ***: p ≤ 0.001, and ****: p ≤ 0.0001). |