- Title
-
CRIMP: a CRISPR/Cas9 insertional mutagenesis protocol and toolkit
- Authors
- Miles, L.B., Calcinotto, V., Oveissi, S., Serrano, R.J., Sonntag, C., Mulia, O., Lee, C., Bryson-Richardson, R.J.
- Source
- Full text @ Nat. Commun.
|
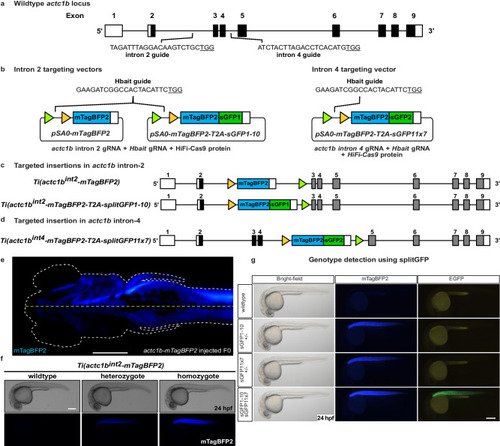
SplitGFP alleles enable visual selection of homozygous mutants. |
|
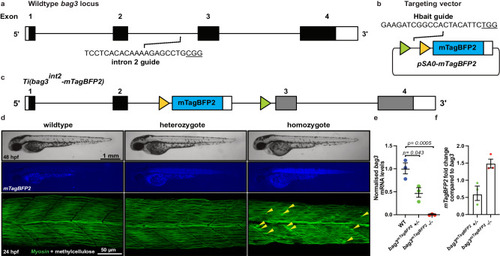
Site-specific integration of targeting vector into |
|
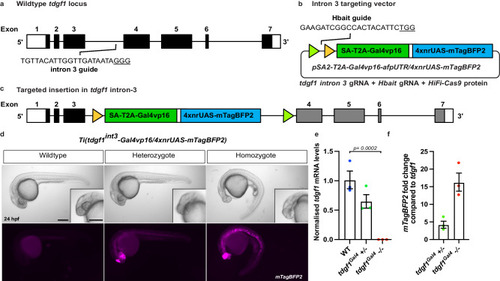
Site-specific integration of targeting vector into |
|
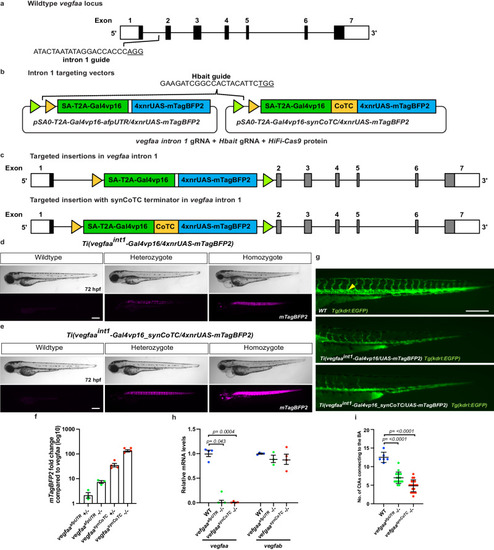
Site-specific integration of targeting vectors into |
|
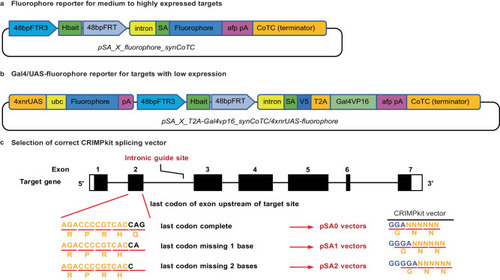
Schematic of CRIMPkit vector design. CRIMPkit vectors contain a guideRNA target site (Hbait) for CRISPR/Cas9 mediated linearisation. The Hbait site is directly upstream of the 3’ region of |