- Title
-
Projective light-sheet microscopy with flexible parameter selection
- Authors
- Chen, B., Chang, B.J., Daetwyler, S., Zhou, F., Sharma, S., Lee, D.M., Nayak, A., Noh, J., Dubrovinski, K., Chen, E.H., Glotzer, M., Fiolka, R.
- Source
- Full text @ Nat. Commun.
|
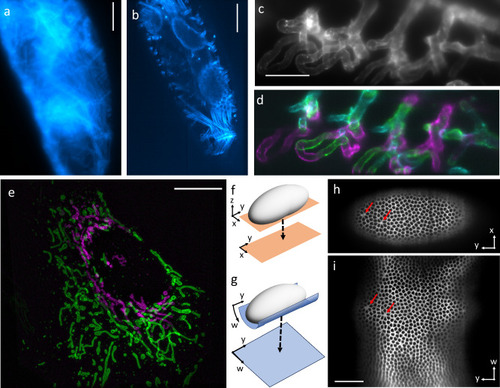
Concept of projections with parameter selection (props). |
|
Variation of projection parameters on different biological samples. |
|
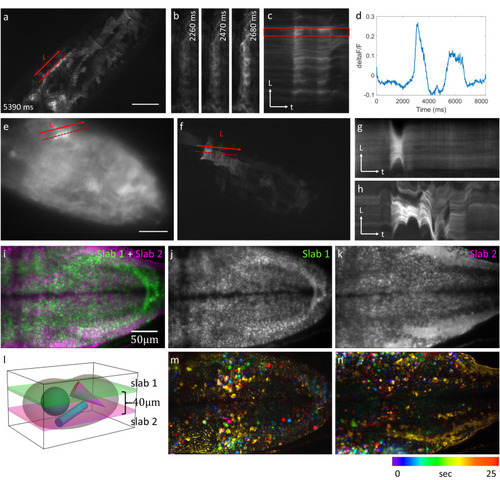
Selective projection imaging of calcium dynamics. |