- Title
-
Variants in UBAP1L lead to autosomal recessive rod-cone and cone-rod dystrophy
- Authors
- Zeitz, C., Navarro, J., Azizzadeh Pormehr, L., Méjécase, C., Neves, L.M., Letellier, C., Condroyer, C., Albadri, S., Amprou, A., Antonio, A., Ben-Yacoub, T., Wohlschlegel, J., Andrieu, C., Serafini, M., Bianco, L., Antropoli, A., Nassisi, M., El Shamieh, S., Chantot-Bastaraud, S., Mohand-Saïd, S., Smirnov, V., Sahel, J.A., Del Bene, F., Audo, I.
- Source
- Full text @ Genet. Med.
|
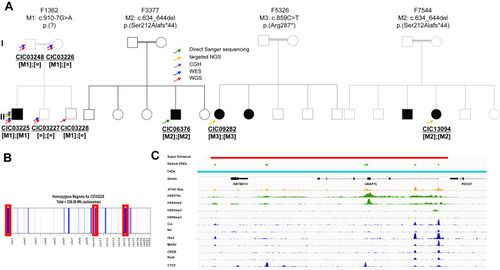
Families with autosomal recessive RCD and CRD/CD and pathogenic variants in UBAP1L. A. Pedigrees of the 4 families (F1362, F3377, F5326, and F7544) with variants in UBAP1L cosegregating with the phenotype in all available family members. The arrows indicate the genetic screening performed on each subject. DNA from the family members of family F3377, F5326, and F7544 were unavailable, but because of the reported consanguinity, we presumed that M2 and M3 occurred homozygous in the affected index cases. B. Homozygosity mapping using WGS data identified 3 large (>20 Mb) homozygous regions in the affected index individual CIC03225 of F1362 with sizes of 27.7, 20.9, and 25.8 Mb mapping to chromosome 1, 10, and 15, respectively. C. Super Enhancer (red), retinal cis-regulatory elements (light green), topologically associated domain (teal), chromatin accessibility (orange), histone modifications (green), and transcription factors residency (blue) for the UBAP1L locus. Chromatin accessibility has been defined by ATAC-seq from post-mortem adult human retina. Histone modifications and transcription factor residency have been defined by ChIP-seq from post-mortem adult human retina. Super enhancer and retinal cis-regulator elements have been calculated from previously described experimental data.21,22 |
|
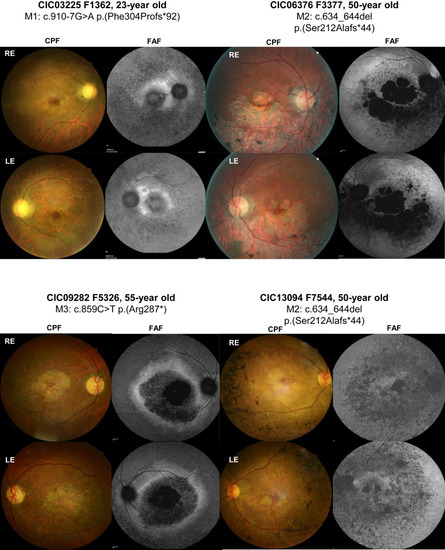
Retinal images of index cases carrying homozygous variants in UBAP1L (CIC03225, CIC06376, CIC09282, and CIC13094). Color fundus photographs (CFP) and fundus autofluorescence imaging (FAF) of the right and left eye (RE and LE, respectively). CIC03225’s fundus examination revealed typical signs of RCD with pale optic discs, narrowed retinal vessels, and central macular atrophy and a loss of autofluorescence in the periphery, as well as in the macular region. CIC06376 showed central macular atrophy expending into the mid periphery predominantly in the inferior sector and a loss of autofluorescence in the atrophic areas surrounded by an area of heterogeneous autofluorescence expanding in the mid periphery. CIC09282 had pale optic discs, narrowed retinal vessels, and atrophic changes in the macular area with a loss of central autofluorescence surrounded by a ring of increased autofluorescence. CIC13094 had central macular atrophy and speckled appearance of autofluorescence at the posterior pole and mid periphery. |
|
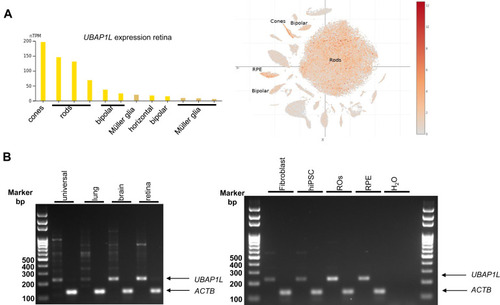
Expression analysis of UBAP1L mRNA in human tissues by RT-PCR experiments. A. Expression analyses using databases revealed that the human UBAP1L gene is expressed in the eye and more specifically in RPE cells and rod and cone photoreceptors with a low expression in bipolar cells. B. UBAP1L was found to be expressed in human universal embryonal tissue and lung but, more importantly, in brain, retina, fibroblasts, hiPSCs, retinal organoids (ROs), and RPE cells differentiated from hiPSCs. Other unspecific transcripts appeared in universal tissue and lung. |
|
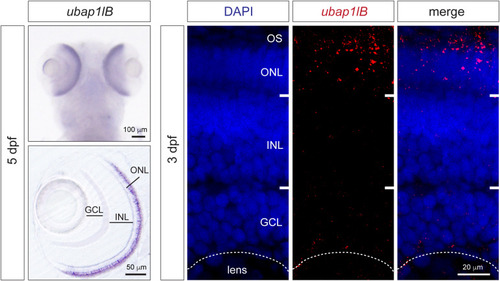
Expression analysis of ubap1l in zebrafish. In situ hybridization for ubap1lb in zebrafish larvae 5 days after fertilization (left). The dorsal view is on the top and retina vibratome section is on the bottom. Expression can be detected in the retina in the photoreceptor layer. On the right, fluorescent HCR in situ hybridization on 3 days after fertilization larvae confirms ubap1lb mRNA expression in the OS and ONL where photoreceptors are located. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, Ganglion cell layer; OS, outer segment. EXPRESSION / LABELING:
|
|
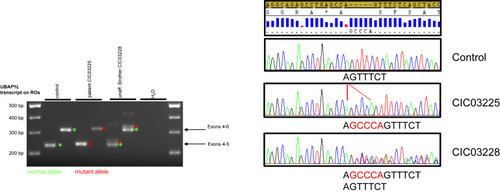
Mis-splicing in ROs of an affected individual lead to a longer protein. RT-PCR experiments performed on ROs at 150 days from the affected individual, CIC03225, revealed a 5-nucleotide insertion, whereas the unaffected heterozygous brother, CIC03228, revealed both the 5-nucleotide insertion and the reference sequence on transcript level, as confirmed by Sanger sequencing. These findings confirmed the in silico prediction for mis-splicing of the c.910-7G>A in UBAP1L, leading most likely to a longer abnormal protein p.(Phe304Profs∗92). |
|
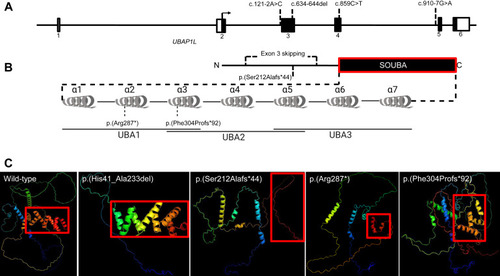
Gene structure, prediction of domains and 3D-modeling of the unaffected and altered UBAP1L protein. A. Schematic representation of UBAP1L gene structure with the positions of identified variants. B. Schematic representation of UBAP1L protein structure with the positions of identified variants. The SOUBA domain structure is detailed showing the composition of UBAs and α-helices. C. AlphaFold 3D structure predictions of UBAP1L wild-type (UniprotKB: F5GYI3), p.(His41_Ala233del), p.(Ser212Alafs∗44), p.(Arg287∗), and p.(Phe304Profs∗92). The color scheme is a gradient that goes from N terminus (blue) to C terminus (red). |