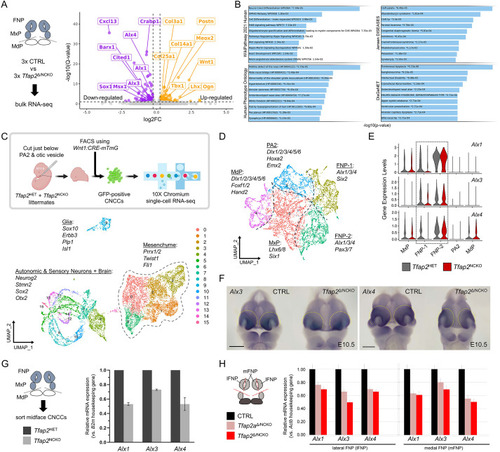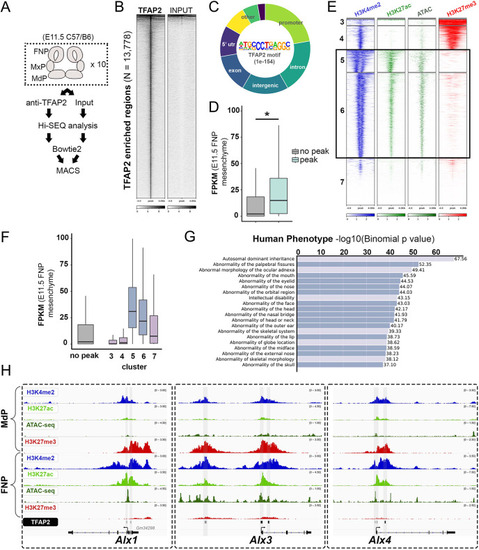
ChIP-seq profiling identifies TFAP2 paralogs directly bind Alx1, Alx3 and Alx4 regulatory elements. (A) Cartoon workflow for anti-TFAP2 ChIP-seq of wild-type E11.5 C57BL/6 facial prominence tissues. (B) Density heatmap displaying read-depth at the 13,778 TFAP2 ChIP-seq ‘peaks’ (y-axis) relative to non-immunoprecipitated input. (C) Hollow pie chart summarizing the distribution of TFAP2 ‘peaks’ throughout the genome, relative to key features (e.g. promoters, introns, etc.). The TFAP2 motif, along with the significance of its enrichment in all TFAP2 ‘peaks’, is displayed in the center. (D) Box-and-whisker graph plotting the FPKM values of 15,842 genes expressed in the E11.5 frontonasal prominence (FNP) mesenchyme (Hooper et al., 2020), with or without an associated TFAP2 peak (*P=2.60e-256, unpaired Wilcoxon test). (E) Density heatmaps for anti-H3K4me2 (blue), anti-H3K27ac (green), ATAC-seq (forest green) and anti-H3K27me3 (red) profiles of E10.5 FNP CNCCs (Minoux et al., 2017) at the TFAP2-positive coordinates, divided by k-means clustering. (F) Box-and-whisker graph, as in D, but with further partitioning the ‘peak’ group based on k-means clusters from E. Boxes represent the 1st and 3rd quartiles; whiskers represent the spread of values, excluding outliers (i.e. within 1.5× of the inter-quartile range). (G) GREAT (McLean et al., 2010) pathway analysis of genes associated with cluster 5. ‘Human Phenotype’ terms are listed, with craniofacial-specific terms in darker boxes. (H) IGV browser views of the Alx1, Alx3 and Alx4 loci overlaid with the epigenome signatures (blue, H3K4me2; green, H3K27ac; khaki, ATAC; red, H3K27me3) of E10.5 CNCCs residing in the mandibular prominence (MdP, top four tracks) and FNP (tracks 5-8), as well as the cluster 5-assigned TFAP2 peaks (bottom, gray highlights).
|