- Title
-
Automated, high-throughput quantification of EGFP-expressing neutrophils in zebrafish by machine learning and a highly-parallelized microscope
- Authors
- Efromson, J., Ferrero, G., Bègue, A., Doman, T.J.J., Dugo, C., Barker, A., Saliu, V., Reamey, P., Kim, K., Harfouche, M., Yoder, J.A.
- Source
- Full text @ PLoS One
|
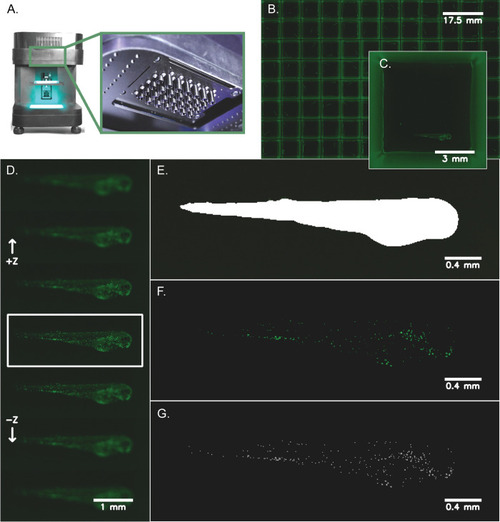
Zebrafish imaging and neutrophil quantification workflow. Transgenic zebrafish larvae ( |
|
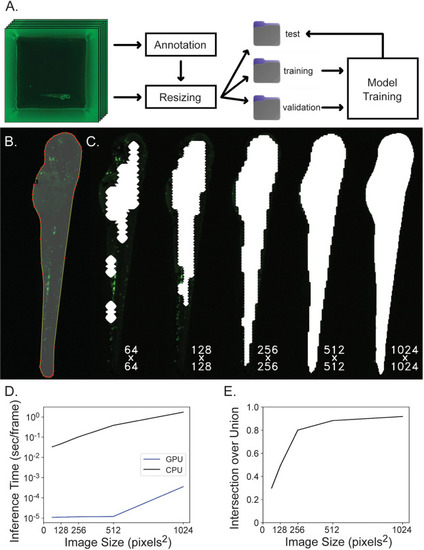
Segmentation network training and evaluation. |
|
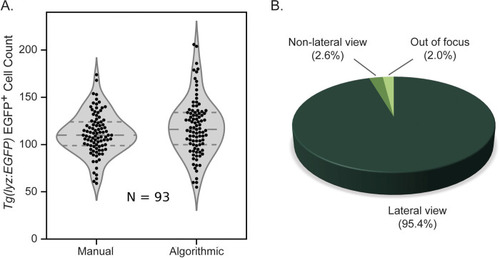
Algorithmic versus manual counting of EGFP+ neutrophils. |
|
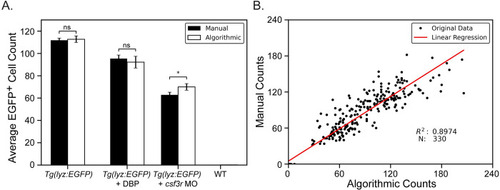
Algorithmic versus manual cell counting for experimental conditions. |